Pan-cancer analysis of ubiquitin-specific protease 7 and its expression changes in the carcinogenesis of scar ulcer
-
摘要:
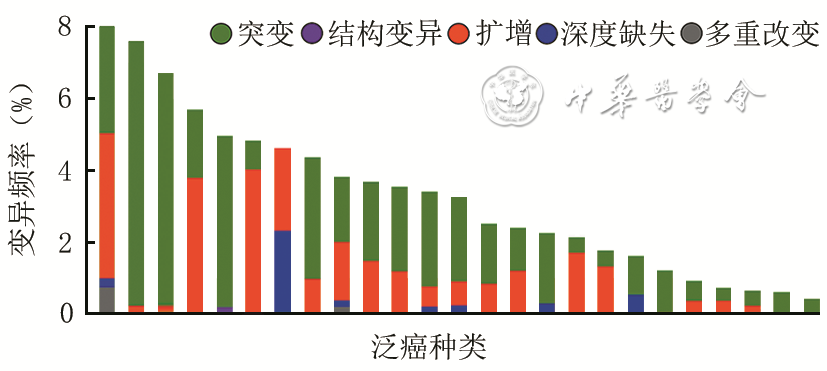
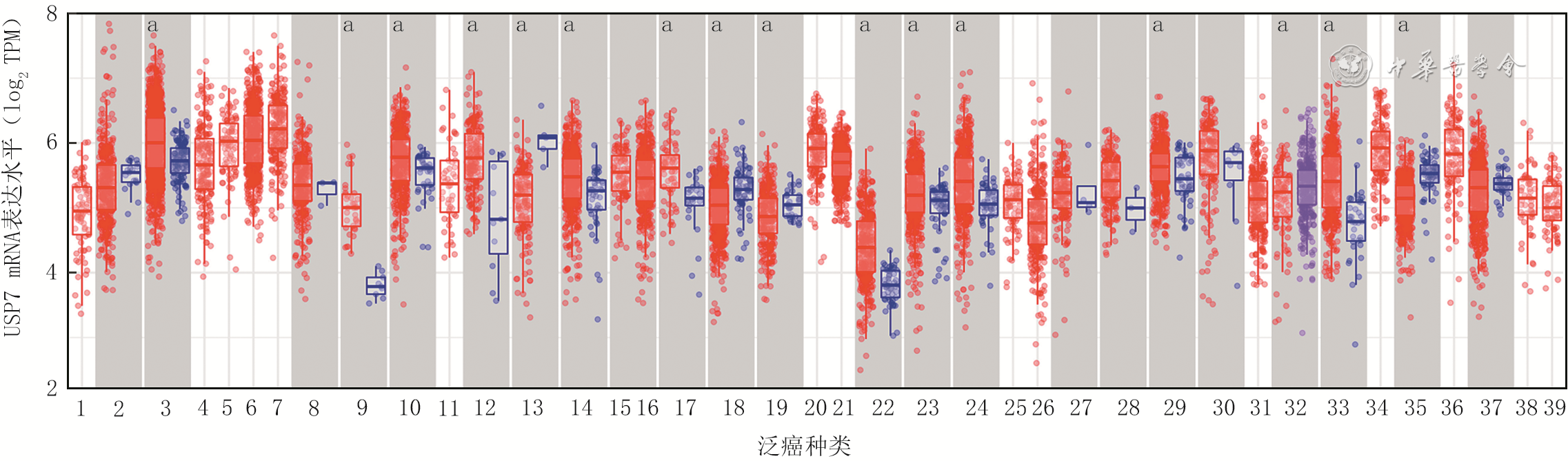
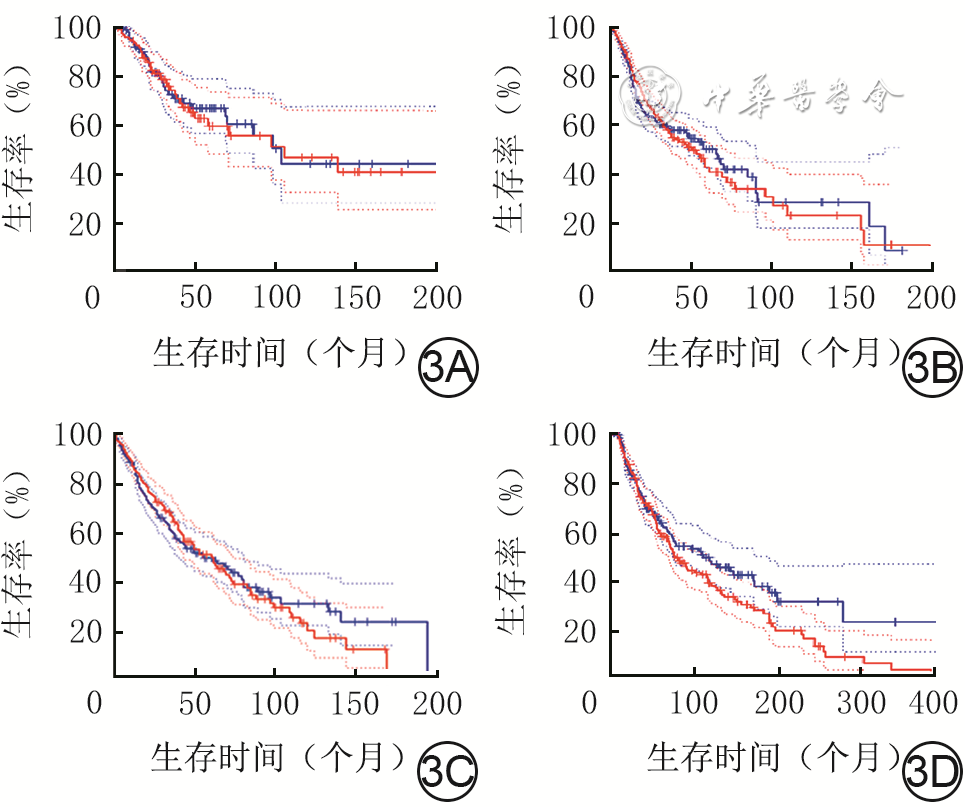
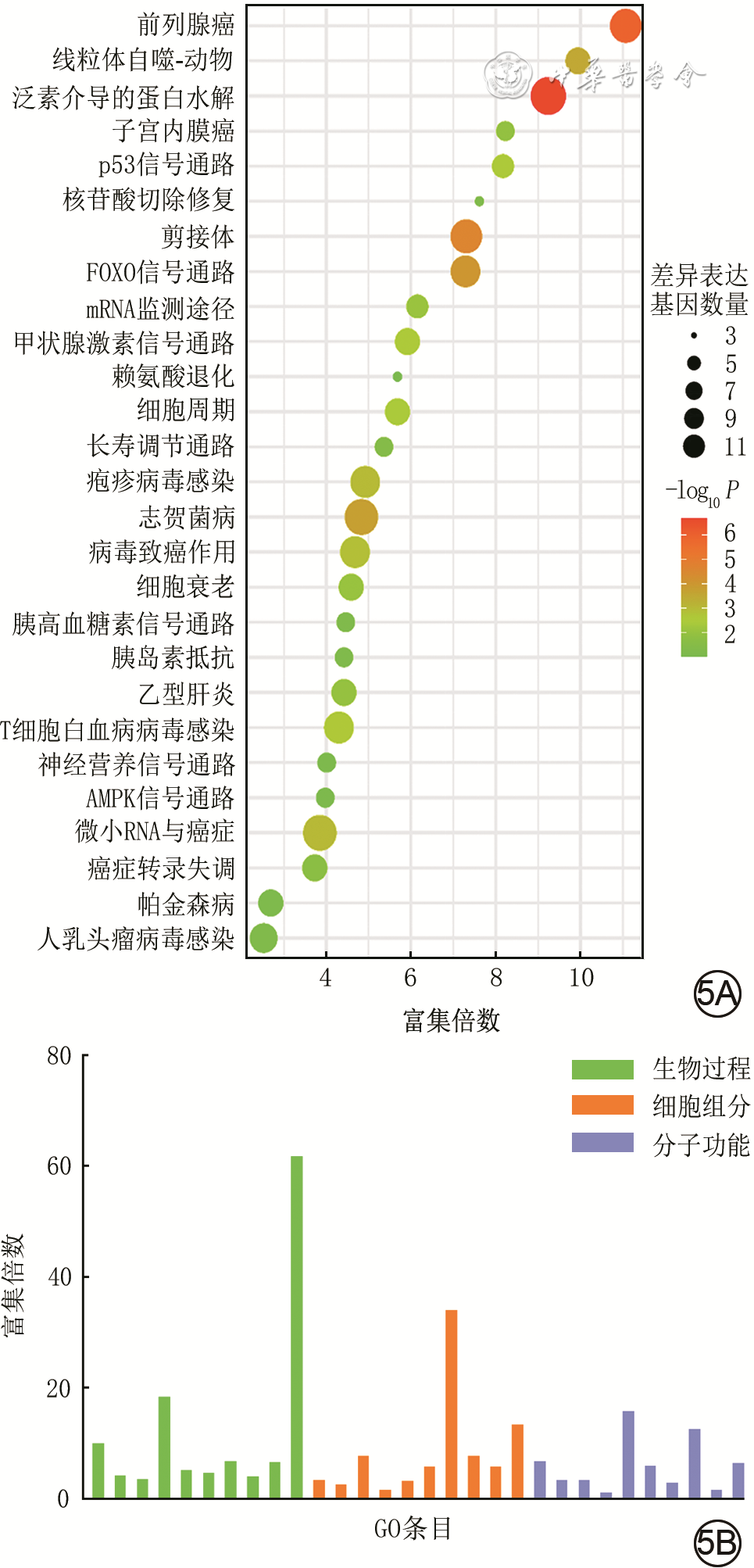
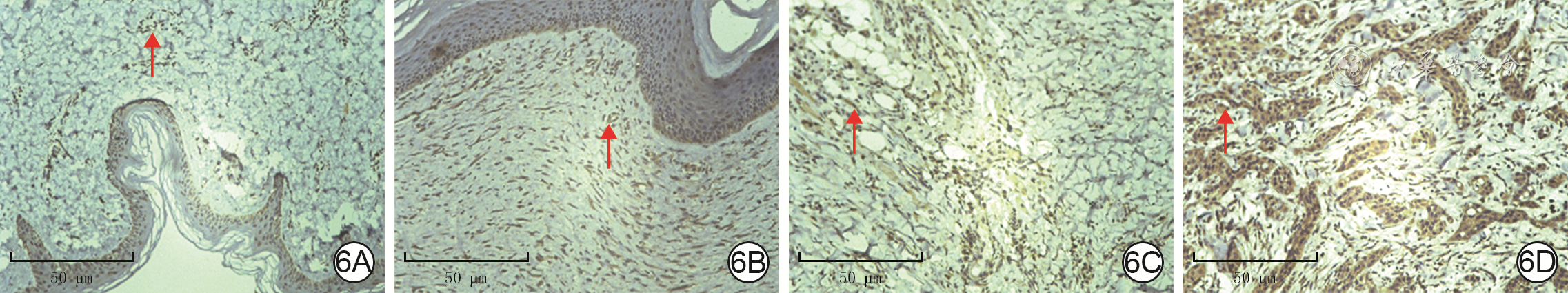
目的 探讨泛素特异性蛋白酶7(USP7)在瘢痕溃疡癌变过程中的生物学作用及其临床意义。 方法 采用回顾性观察性研究结合生物信息学分析方法。从癌症基因组图谱(TCGA)数据库和基因表达综合数据库中获取USP7在肿瘤和/或其对应癌旁正常组织中的RNA表达谱数据,并将RNA测序数据进行log 2转化。通过多维度癌症基因集cBioPortal数据库分析 USP7基因变异情况,并分析其突变位点。通过TIMER 2.0数据库中“差异表达”模块获得TCGA数据库中肿瘤、癌旁正常组织USP7 mRNA表达情况。使用基因表达谱交互分析2(GEPIA2)数据库分析皮肤黑色素瘤(SKCM)、宫颈鳞状细胞癌(CESC)、肺鳞状细胞癌(LUSC)和头颈鳞状细胞癌(HNSC)中高表达USP7患者与低表达USP7患者的生存率并绘制Kaplan-Meier生存曲线。利用Sangerbox数据库分析USP7在泛癌中的表达与微卫星不稳定性(MSI)或肿瘤突变负担(TMB)的相关性。通过GEPIA2数据库中“相关性分析”模块,评估USP7在泛癌中的表达与5种DNA错配修复基因( MLH1、 MSH2、 MSH6、 PMS2和 EPCAM)表达水平和3种必需DNA甲基转移酶(DNMT)——DNMT1、DNMT3A、DNMT3B表达水平的相关性。通过TIMER 2.0数据库中“免疫-基因”模块分析USP7在CESC、HNSC、LUSC和SKCM中表达及其与免疫细胞(B细胞、CD4 +T细胞、CD8 +T细胞、中性粒细胞、巨噬细胞和树突状细胞)浸润的相关性。利用GEPIA2数据库的“相似基因检测”模块获得与USP7表达模式相似且排名前100的蛋白集。将前述蛋白集与利用STRING数据库获得的与USP7有直接物理结合作用且排名前50的蛋白集进行交集分析。通过DAVID数据库对上述2个蛋白集进行京都基因与基因组百科全书(KEGG)和基因本体论(GO)富集分析。收集2018年10月—2022年10月武汉大学附属同仁医院暨武汉市第三医院病理科有相应临床病理特征的正常皮肤、增生性瘢痕、瘢痕溃疡、瘢痕癌的术后标本,采用免疫组织化学法检测组织中USP7表达情况,样本数为6。对数据行Log-rank检验、单因素方差分析及Bonferroni检验。 结果 在泛癌中, USP7的主要基因变异类型为突变和扩增,变异频率(>6%)居前3位的肿瘤依次为膀胱尿路上皮癌、SKCM和子宫内膜癌。 USP7基因在泛癌中的主要突变为错义突变。在突变频率最高的SKCM中,主要突变类型是USP7_ICP0_bdg结构域的错义突变。USP7 mRNA在乳腺浸润癌、胆管癌、结肠癌、食管癌、HNSC、肾嫌色细胞癌、肝细胞肝癌、肺腺癌、LUSC、前列腺癌和胃癌肿瘤组织中的表达均明显高于其对应的癌旁正常组织( P<0.05),在多形成性胶质细胞瘤、肾透明细胞癌、肾乳头状细胞癌和甲状腺癌中的表达均明显低于其对应的癌旁正常组织( P<0.05);此外,USP7 mRNA在SKCM转移组织中的表达远高于其原发肿瘤组织( P<0.05)。生存曲线显示,在CESC、HNSC、LUSC和SKCM中,高表达USP7患者与低表达USP7患者的存活率比较,差异均无统计学意义(Log-rank P>0.05,风险比分别为1.00、0.99、1.00、1.30)。USP7在结肠癌、结直肠癌、胸腺癌、甲状腺癌中的表达均与TMB呈显著负相关(Pearson相关系数分别为-0.26、-0.19、-0.19和-0.11, P<0.05);USP7在神经胶质瘤、CESC、肺腺癌、混合肾癌、LUSC中的表达均与MSI呈显著正相关(Pearson相关系数分别为0.22、0.14、0.15、0.08和0.14, P<0.05),在结肠癌、结直肠癌、乳腺浸润癌、前列腺癌、HNSC、甲状腺癌和弥漫性大B细胞淋巴瘤中的表达均与MSI呈显著负相关(Pearson相关系数分别为-0.31、-0.27、-0.13、-0.19、-0.16、-0.18和-0.53, P<0.05)。USP7在CESC中的表达与MSH2和MSH6的表达均呈明显正相关(Spearman相关系数分别为0.51和0.44, P<0.05),在HNSC中的表达与EPCAM、MLH1、MSH2、MSH6、PMS2的表达均呈明显正相关(Spearman相关系数分别为0.39、0.14、0.49、0.54和0.41, P<0.05),在LUSC中的表达与EPCAM、MSH2、MSH6和PMS2的表达均呈明显正相关(Spearman相关系数分别为0.20、0.36、0.40和0.34, P<0.05),在SKCM中的表达与EPCAM、MLH1、MSH2、MSH6和PMS2的表达均呈明显正相关(Spearman相关系数分别为0.11、0.33、0.42、0.55和0.34, P<0.05);USP7在CESC、HNSC、LUSC和SKCM中的表达与DNMT1、DNMT3A和DNMT3B的表达均呈显著正相关(Spearman相关系数分别为0.42、0.34和0.22,0.45、0.52和0.22,0.36、0.36和0.22,0.38、0.46和0.21, P<0.05)。USP7在CESC、HNSC、LUSC和SKCM中的表达仅与CD4 +T细胞浸润呈显著正相关(Partial相关系数分别为0.14、0.22、0.13、0.16, P<0.05)。与USP7表达模式相似且排名前100的蛋白集中,排名前5的蛋白依次是C16orf72、BCLAF1、UBN、GSPT1、ERI2(Spearman相关系数分别为0.83、0.74、0.73、0.73和0.72, P值均<0.05)。与USP7有直接物理结合作用且排名前50的蛋白集与前述蛋白集的交集仅有1个蛋白,即为甲状腺激素受体相互作用因子12。KEGG富集分析显示,USP7相关基因涉及细胞周期、剪接体、细胞衰老和p53信号通路等。GO富集分析显示,USP7相关基因涉及转录调控、蛋白质泛素化、DNA修复和细胞质模式识别受体信号通路等。临床样本分析显示,USP7在增生性瘢痕(0.35±0.05)、瘢痕溃疡(0.43±0.04)和瘢痕癌(0.61±0.03)中的表达均明显高于正常皮肤(0.18±0.04), P<0.05。 结论 USP7可能为临床瘢痕溃疡癌变恶化的生物标志物。 Abstract:Objective To explore the biological role and clinical significance of ubiquitin-specific protease 7 (USP7) in the carcinogenesis of scar ulcer. Methods A retrospective observational study combined with bioinformatics analysis was used. The RNA expression profile data of USP7 in tumor and/or its corresponding paracancular normal tissue were obtained from The Cancer Genome Atlas (TCGA) database and the Gene Expression Omnibus database, and the RNA sequencing data were transformed by log 2. The variations of USP7 gene were analyzed by cBioPortal database. The USP7 mRNA expression in tumor and adjacent normal tissue in TCGA database were obtained by using the "Gene_DE" module in TIMER 2.0 database. The survival rates of patients with high and low USP7 expression in cutaneous melanoma (SKCM), cervical squamous cell carcinoma (CESC), lung squamous cell carcinoma (LUSC), and head and neck squamous cell carcinoma (HNSC) were analyzed using the Gene Expression Profile Interactive Analysis 2 (GEPIA2) database, and the Kaplan-Meier survival curves were drawn. Sangerbox database was used to analyze the correlation of USP7 expression in pan-cancer with microsatellite instability (MSI) or tumor mutation burden (TMB) pan-cancer. Through the "correlation analysis" module in the GEPIA2 database, the correlation of USP7 expression in pan-cancer with the expression levels of five DNA mismatch repair genes ( MLH1, MSH2, MSH6, PMS2, and EPCAM) and three essential DNA methyltransferases (DNMT)--DNMT1, DNMT3A, and DNMT3B were evaluated. The USP7 expression in CESC, HNSC, LUSC, and SKCM and its correlation with infiltration of immune cells (B cells, CD4 + T cells, CD8 + T cells, neutrophils, macrophages, and dendritic cells) were analyzed by the "Immune-Gene" module in TIMER 2.0 database. The "Similar Genes Detection" module of GEPIA2 database was used to obtain the top 100 protein sets with similar expression patterns to USP7. Intersection analysis was performed between the aforementioned protein sets and the top 50 protein sets that were directly physically bound to USP7 obtained by using the STRING database. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analysis were performed for the two protein sets mentioned above using the DAVID database. The samples of normal skin, hypertrophic scar, scar ulcer, and scar carcinoma with corresponding clinicopathologic features were collected from the Department of Pathology of Tongren Hospital of Wuhan University & Wuhan Third Hospital from October 2018 to October 2022, and the USP7 expression in tissue was detected by immunohistochemical method, with the number of samples of 6. Data were statistically analyzed with Log-rank test, one-way analysis of variance, and Bonferroni test. Results In pan-cancer, the main gene variations of USP7 were mutation and amplification, and the top 3 tumors with the highest variation frequency (>6%) were bladder urothelial carcinoma, SKCM, and endometrial carcinoma. The main mutation of USP7 gene in pan-cancer was missense mutation. In SKCM with the highest mutation frequency, the main type of mutation was missense mutation in USP7_ICP0_bdg domain. USP7 mRNA expression in breast invasive carcinoma, bile duct carcinoma, colon carcinoma, esophageal carcinoma, HNSC, renal chromophobe cell carcinoma, hepatocellular carcinoma, lung adenocarcinoma, LUSC, prostate carcinoma, and gastric carcinoma was significantly higher than that in corresponding paracancer normal tissue ( P<0.05). USP7 mRNA expression in glioblastoma multiforme, renal clear cell carcinoma, renal papillary cell carcinoma, and thyroid carcinoma was significantly lower than that in corresponding paracancular normal tissue ( P<0.05). In addition, USP7 mRNA expression in SKCM metastases was much higher than that in primary tumor tissue ( P<0.05). Survival curves showed no significant difference in survival rate between patients with high USP7 expression and patients with low USP7 expression in CESC, HNSC, LUSC, and SKCM (Log-rank P>0.05, with hazard ratios of 1.00, 0.99, 1.00, and 1.30, respectively). USP7 expression in colon cancer, colorectal cancer, thymic cancer, and thyroid cancer was negatively correlated with TMB (with Pearson correlation coefficients of -0.26, -0.19, -0.19, and 0.11, respectively, P<0.05). USP7 expression in glioma, CESC, lung adenocarcinoma, mixed renal carcinoma, and LUSC was positively correlated with MSI expression (with Pearson correlation coefficients of 0.22, 0.14, 0.15, 0.08, and 0.14, respectively, P<0.05), and USP7 expression in colon cancer, colorectal cancer, invasive breast cancer, prostate cancer, HNSC, thyroid cancer, and diffuse large B-cell lymphoma were significantly negatively correlated with MSI expression (with Pearson correlation coefficients of -0.31, -0.27, -0.13, -0.19, -0.16, -0.18, and -0.53, respectively, P<0.05). The expression of USP7 in CESC was positively correlated with that of both MSH2 and MSH6 (with Spearman correlation coefficients of 0.51 and 0.44, respectively, P<0.05), and the expression of USP7 in HNSC was positively correlated with the expression of EPCAM, MLH1, MSH2, MSH6, and PMS2 (with Spearman correlation coefficients of 0.39, 0.14, 0.49, 0.54, and 0.41, respectively, P<0.05), and the expression of USP7 in LUSC was positively correlated with the expression of EPCAM, MSH2, MSH6, and PMS2 (with Spearman correlation coefficients of 0.20, 0.36, 0.40, and 0.34, respectively, P<0.05), and the expression of USP7 in SKCM was positively correlated with the expression of EPCAM, MLH1, MSH2, MSH6, and PMS2 (with Spearman correlation coefficients of 0.11, 0.33, 0.42, 0.55, and 0.34, respectively, P<0.05). The expression of USP7 in CESC, HNSC, LUSC, and SKCM was significantly positively correlated with the expression of DNMT1, DNMT3A, and DNMT3B (with Spearman correlation coefficients of 0.42, 0.34, 0.22, 0.45, 0.52, 0.22, 0.36, 0.36, 0.22, 0.38, 0.46, and 0.21, respectively, P<0.05). The expression of USP7 in CESC, HNSC, LUSC, and SKCM was positively correlated with CD4 + T cell infiltration (with Partial correlation coefficients of 0.14, 0.22, 0.13, and 0.16, respectively, P<0.05). Being similar to the pattern of USP7 expression and ranked among top 100 protein sets, the top 5 proteins were C16orf72, BCLAF1, UBN, GSPT1, ERI2 (with Spearman correlation coefficients of 0.83, 0.74, 0.73, and 0.72, respectively, all P values<0.05). The top 50 protein sets that directly physically bind to USP7 overlapped with the aforementioned protein set by only one protein, thyroid hormone receptor interaction factor 12. KEGG enrichment analysis showed that USP7 related genes were involved in cell cycle, spliceosome, cell senescence, and p53 signal pathway. GO enrichment analysis showed that USP7 related genes were involved in transcriptional regulation, protein ubiquitination, DNA repair, and cytoplasmic pattern recognition receptor signal pathways. Analysis of clinical samples showed that USP7 expression was significantly higher in hypertrophic scars (0.35±0.05), scar ulcers (0.43±0.04), and scar cancers (0.61±0.03) than in normal skin (0.18±0.04), P<0.05. Conclusions USP7 may be a clinical biomarker for the progression of cicatricial ulcer cancer. -
Key words:
- Skin neoplasms /
- Cicatrix /
- Ubiquitin-specific protease 7 /
- Pan-cancer analysis /
- Scar ulcer /
- Cicatricial carcinoma /
- Wound repair
-
章思语:酝酿、设计和实施研究,采集、分析、解释数据,撰写文章;阮晶晶,金冬梅:采集、分析、解释数据,研究指导,对文章的知识性内容作批判性审阅;陈诺:采集数据,对文章的知识性内容作批判性审阅;谢卫国:分析、解释数据,对文章的知识性内容作批判性审阅;阮琼芳:对文章的知识性内容作批判性审阅,经费支持,研究指导所有作者均声明不存在利益冲突
-
参考文献
(23) [1] ShpichkaA, ButnaruD, BezrukovEA, et al. Skin tissue regeneration for burn injury[J]. Stem Cell Res Ther, 2019,10(1):94. DOI: 10.1186/s13287-019-1203-3. [2] BazalińskiD, Przybek-MitaJ, BarańskaB, et al. Marjolin's ulcer in chronic wounds - review of available literature[J]. Contemp Oncol (Pozn), 2017,21(3):197-202. DOI: 10.5114/wo.2017.70109. [3] ShenW, ZhangZ, MaJ, et al. The ubiquitin proteasome system and skin fibrosis[J]. Mol Diagn Ther, 2021,25(1):29-40. DOI: 10.1007/s40291-020-00509-z. [4] El AyadiA, JayJW, PrasaiA. Current approaches targeting the wound healing phases to attenuate fibrosis and scarring[J]. Int J Mol Sci, 2020,21(3):1105.DOI: 10.3390/ijms21031105. [5] TorisevaM, KähäriVM. Proteinases in cutaneous wound healing[J]. Cell Mol Life Sci, 2009,66(2):203-224. DOI: 10.1007/s00018-008-8388-4. [6] NininahazweL, LiuB, HeC, et al. The emerging nature of Ubiquitin-specific protease 7 (USP7): a new target in cancer therapy[J]. Drug Discov Today, 2021,26(2):490-502. DOI: 10.1016/j.drudis.2020.10.028. [7] WangZ, KangW, YouY, et al. USP7: novel drug target in cancer therapy[J]. Front Pharmacol, 2019,10:427. DOI: 10.3389/fphar.2019.00427. [8] GuoL, ChenL, BiS, et al. PTEN inhibits proliferation and functions of hypertrophic scar fibroblasts[J]. Mol Cell Biochem, 2012,361(1/2):161-168. DOI: 10.1007/s11010-011-1100-2. [9] LiS, DingX, YanX, et al. ceAF ameliorates diabetic wound healing by alleviating inflammation and oxidative stress via TLR4/NF-κB and Nrf2 pathways[J]. J Diabetes Res, 2023,2023:2422303. DOI: 10.1155/2023/2422303. [10] MiaoC, LiY, ZhangX. The functions of FoxO transcription factors in epithelial wound healing[J]. Australas J Dermatol, 2019,60(2):105-109. DOI: 10.1111/ajd.12952. [11] HeT, BaiX, JingJ, et al. Notch signal deficiency alleviates hypertrophic scar formation after wound healing through the inhibition of inflammation[J]. Arch Biochem Biophys, 2020,682:108286. DOI: 10.1016/j.abb.2020.108286. [12] GannonHS, DonehowerLA, LyleS, et al. Mdm2-p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin[J]. Dev Biol, 2011,353(1):1-9. DOI: 10.1016/j.ydbio.2011.02.007. [13] GotoK, IshikawaM, AizawaD, et al. Nuclear β-catenin immunoexpression in scars[J]. J Cutan Pathol, 2021,48(1):18-23. DOI: 10.1111/cup.13806. [14] ChenZY, YuXF, HuangJQ, et al. The mechanisms of β‐catenin on keloid fibroblast cells proliferation and apoptosis[J]. Eur Rev Med Pharmacol Sci, 2018,22(4):888-895. DOI: 10.26355/eurrev_201802_14366. [15] ChenF, WendlMC, WyczalkowskiMA, et al. Moving pan-cancer studies from basic research toward the clinic[J]. Nat Cancer, 2021,2(9):879-890. DOI: 10.1038/s43018-021-00250-4. [16] ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes[J]. Nature, 2020,578(7793):82-93. DOI: 10.1038/s41586-020-1969-6. [17] HussainI, ul RehmanS, AfrozeD, et al. Mutational spectrum of conserved regions of TP53 and PTEN genes in Kangri cancer (of the skin) in the Kashmiri population[J]. Mutat Res, 2009,676(1/2):5-10. DOI: 10.1016/j.mrgentox.2009.02.011. [18] TengY, FanY, MaJ, et al. The PI3K/Akt pathway: emerging roles in skin homeostasis and a group of non-malignant skin disorders[J]. Cells, 2021,10(5):1219.DOI: 10.3390/cells10051219. [19] ChungJY, ChanMK, LiJS, et al. TGF-β signaling: from tissue fibrosis to tumor microenvironment[J]. Int J Mol Sci, 2021,22(14):7575.DOI: 10.3390/ijms22147575. [20] InnellaG, BonoraE, NeriI, et al. PTEN hamartoma tumor syndrome: skin manifestations and insights into their molecular pathogenesis[J]. Front Med (Lausanne), 2021,8:688105. DOI: 10.3389/fmed.2021.688105. [21] MorottiA, PanuzzoC, CrivellaroS, et al. BCR-ABL disrupts PTEN nuclear-cytoplasmic shuttling through phosphorylation-dependent activation of HAUSP[J]. Leukemia, 2014,28(6):1326-1333. DOI: 10.1038/leu.2013.370. [22] LuJ, ZhaoH, YuC, et al. Targeting Ubiquitin-specific protease 7 (USP7) in cancer: a new insight to overcome drug resistance[J]. Front Pharmacol, 2021,12:648491. DOI: 10.3389/fphar.2021.648491. [23] BrooksCL, LiM, HuM, et al. The p53--Mdm2--HAUSP complex is involved in p53 stabilization by HAUSP[J]. Oncogene, 2007,26(51):7262-7266. DOI: 10.1038/sj.onc.1210531. -
2 癌症基因组图谱数据库中泛素特异性蛋白酶7(USP7)mRNA表达水平分析的箱式图
注:TPM为样本中平均每一百万个转录本中对应特定基因或转录本的数量;横坐标上从左到右的数字依次代表肾上腺皮质癌、膀胱尿路上皮癌、乳腺浸润癌、乳腺浸润基底癌、人表皮生长因子受体阳性乳腺浸润癌、LumA乳腺浸润癌、LumB乳腺浸润癌、宫颈癌、胆管癌、结肠癌、弥漫性大B细胞淋巴瘤、食管癌、多形成性胶质细胞瘤、头颈鳞状细胞癌、人乳头瘤病毒阳性头颈鳞状细胞癌、人乳头瘤病毒阴性头颈鳞状细胞癌、肾嫌色细胞癌、肾透明细胞癌、肾乳头状细胞癌、急性髓细胞样白血病、脑低级别胶质瘤、肝细胞肝癌、肺腺癌、肺鳞状细胞癌、间皮瘤、卵巢浆液性囊腺癌、胰腺癌、嗜铬细胞瘤、前列腺癌、直肠腺癌、肉瘤、皮肤黑色素瘤、胃癌、睾丸癌、甲状腺癌、胸腺癌、子宫内膜癌、子宫肉瘤、葡萄膜黑色素瘤;红色代表肿瘤组织,蓝色代表正常对照组织,紫色代表癌转移组织;有对应正常组织/癌转移组织对照的癌种类背景色为灰色;与正常组织/癌转移组织相比,aP<0.05
4 泛癌中泛素特异性蛋白酶7(USP7)表达与肿瘤相关性的热图分析。4A.USP7表达与DNA错配修复基因表达的相关性分析;4B.USP7表达与DNA甲基转移酶基因表达的相关性分析
注:DNMT为DNA甲基转移酶;PMS2、MSH6、MSH2、MLH1和EPCAM指DNA错配修复基因对应的表达;图4A和图4B中横坐标上癌组织种类及排序相同,从左到右均依次为肾上腺皮质癌、膀胱尿路上皮癌、乳腺浸润癌、乳腺浸润基底癌、人表皮生长因子受体2阳性乳腺浸润癌、LumA乳腺浸润癌、LumB乳腺浸润癌、宫颈鳞状细胞癌、胆管癌、结肠癌、弥漫性大B细胞淋巴瘤、食管癌、多形成性胶质细胞瘤、头颈鳞状细胞癌、人乳头瘤病毒阳性头颈鳞状细胞癌、人乳头瘤病毒阴性头颈鳞状细胞癌、肾嫌色细胞癌、肾透明细胞癌、肾乳头状细胞癌、脑低级别胶质瘤、肝细胞肝癌、肺腺癌、肺鳞状细胞癌、间皮瘤、卵巢浆液性囊腺癌、胰腺癌、嗜铬细胞瘤、前列腺癌、直肠腺癌、肉瘤、皮肤黑色素瘤、转移性皮肤黑色素瘤、皮肤黑色素瘤原位癌、胃癌、睾丸癌、甲状腺癌、胸腺癌、子宫内膜癌、子宫肉瘤、葡萄膜黑色素瘤
5 泛癌中泛素特异性蛋白酶7相关信号通路的京都基因与基因组百科全书(KEGG)和基因本体论(GO)富集分析。5A.KEGG富集分析;5B.GO富集分析
注:FOX为叉头样转录因子,AMPK为腺苷一磷酸活化蛋白激酶;图5B横坐标从左到右依次为生物过程:蛋白质去泛素化、RNA PolⅡ启动子转录的负调控、RNA PolⅡ启动子转录的正调控、转录偶联核苷酸切除修复、病毒过程、蛋白质泛素化、泛素依赖性蛋白质分解代谢过程、转录正调控DNA模板化、DNA修复、细胞质模式识别受体信号通路,细胞组分:核浆、核、核斑点、胞质、染色质、核体、PcG蛋白复合物、核糖核蛋白复合物、染色体、转录抑制因子复合物,分子功能:染色质绑定、核糖核酸绑定、DNA结合、蛋白结合、启动子特异性染色质结合、泛素蛋白转移酶活性、锌离子结合、p53绑定、金属离子结合、组蛋白绑定
-
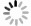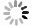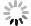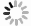







 下载:
下载: