Weighted gene co-expression network analysis of methylated genes in burn scar tissue
-
摘要:
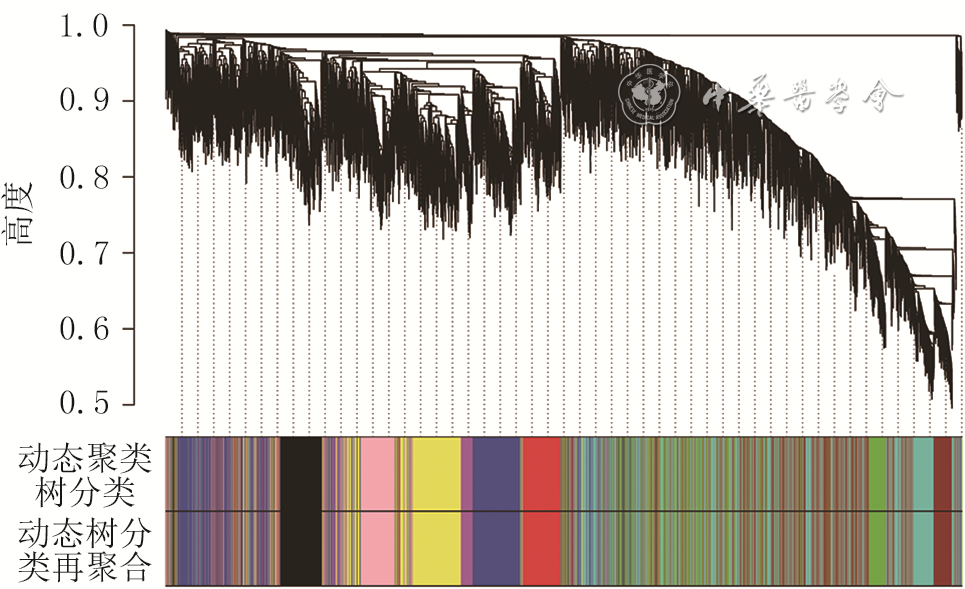
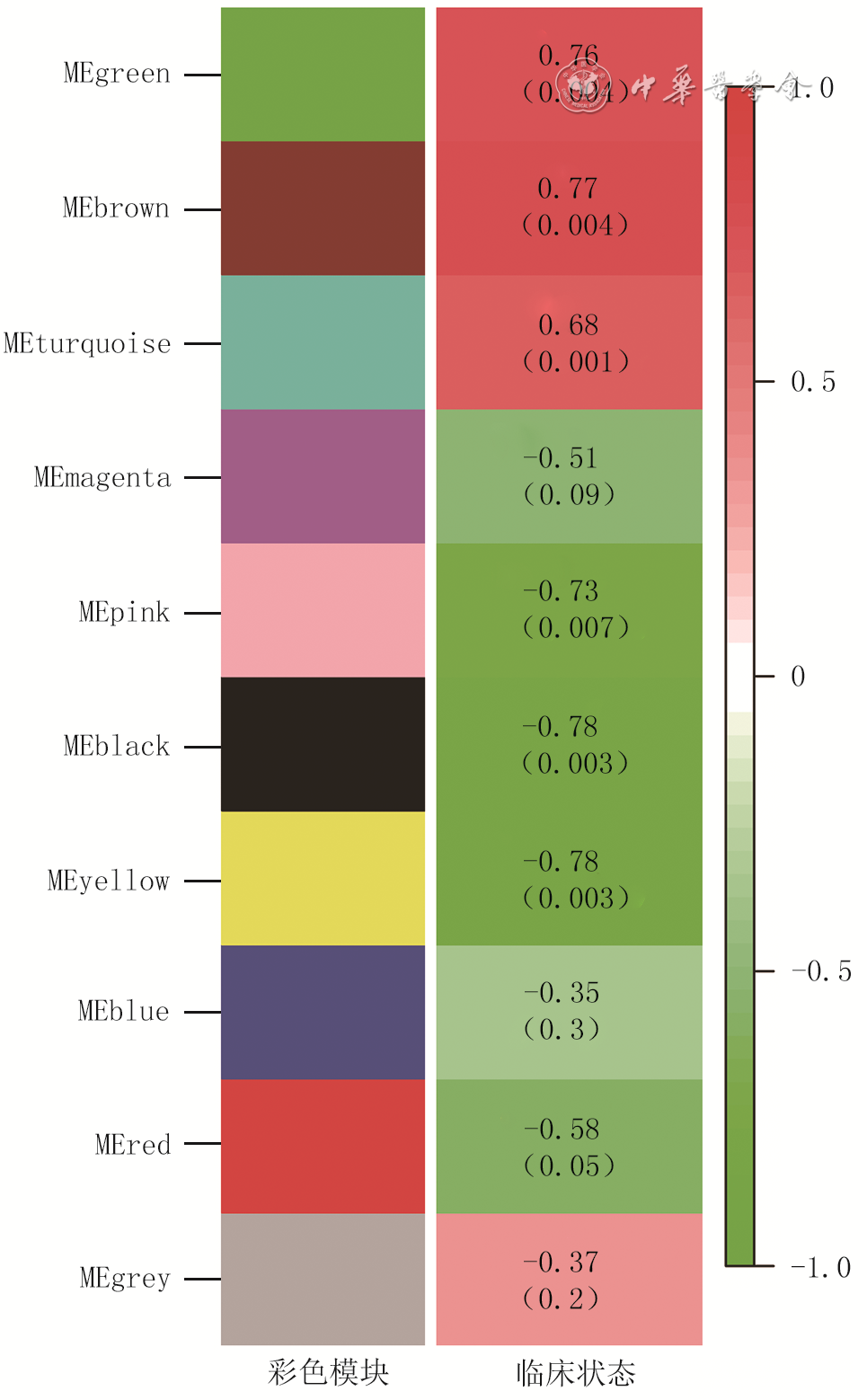
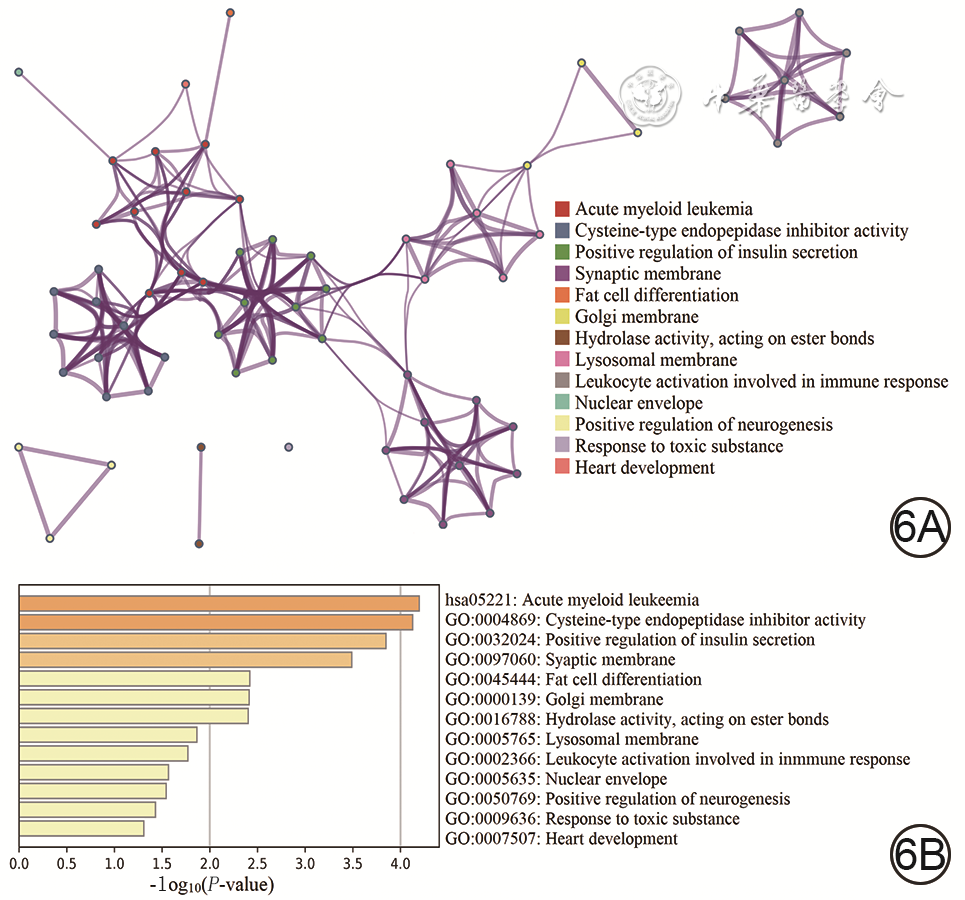
目的 采用加权基因共表达网络分析(WGCNA)探讨烧伤后瘢痕组织内甲基化基因,发掘瘢痕形成过程中的分子标志物及治疗靶点。 方法 采用观察性研究方法。从美国国家生物技术信息中心基因表达综合数据库筛选出同一批次分别进行了mRNA测序和甲基化测序的GSE136906和GSE137134数据集(数据集的样本数均为6),并利用数据集GSE108110(样本数为9)纳入支持向量机及建模分析。采用“Limma”软件包对烧伤后瘢痕和正常组织之间的差异表达基因和差异甲基化基因进行鉴定。使用WGCNA筛选和瘢痕组织临床特征相关性强且基因数目多的模块。对模块内的基因进行功能富集分析并寻找模块内异常甲基化状态的基因,使用受试者操作特征(ROC)曲线判断异常甲基化基因对瘢痕的诊断效能,并使用支持向量机模型进行验证。 结果 共鉴定出10个共表达基因模块,棕色模块与烧伤后瘢痕组织形成相关性高且基因数目多。棕色模块内的基因主要富集在雄激素受体信号通路的调控、细胞因子-细胞因子受体相互作用、胰岛素分泌的正调节等方面。棕色模块内显示35个基因甲基化状态异常,ROC曲线(曲线下面积>0.9)与支持向量机模型(准确率为93.3%)表明基因CCR2、LMO7、STEAP4、NNAT和TCF7L2具有良好的瘢痕诊断效能。 结论 CCR2、LMO7、STEAP4、NNAT和TCF7L2可作为烧伤后瘢痕治疗的潜在靶点。 Abstract:Objective To investigate the methylated genes in burn scar tissue by weighted gene co-expression network analysis (WGCNA), and to discover molecular markers and therapeutic targets of scar formation. Methods An observational research method was used. Datasets were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus Database of America. The GSE136906 (n=6) and GSE137134 (n=6) datasets in the same batch were screened out for mRNA sequencing and methylation sequencing respectively, and the dataset GSE108110 (n=9) was incorporated into support vector machine and modeling analysis. The Limma software package was used to identify the differentially expressed genes and differentially methylated genes between scar tissue after burn and normal tissue. WGCNA was used to select the module with strong correlation with clinical features of scar tissue and large number of genes. Functional enrichment analysis of the genes in the module was performed to find genes with abnormal methylation. The receiver operating characteristic (ROC) curve was used to judge diagnostic efficacy of genes with abnormal methylation for scar, and support vector machine (SVM) was used to verify. Results A total of 10 modules were identified, and the brown module with large number of genes was highly correlated to burn scar tissue formation. The genes in the brown module were mainly concentrated in "regulation of androgen receptor signaling pathway", "cytokine-cytokine receptor interaction", "positive regulation of insulin secretion", and so on. The model showed 35 genes with abnormal methylation status. The ROC curve (area under the curve>0.9) and SVM modeling (accuracy=93.3%) indicated that CCR2, LMO7, STEAP4, NNAT, and TCF7L2 genes had good diagnostic performance for scar. Conclusions CCR2, LMO7, STEAP4, NNAT, and TCF7L2 can be used as potential targets for burn scar treatment. -
Key words:
- Burns /
- Cicatrix /
- Methylation /
- Weighted gene co-expression network /
- CCR2 /
- LMO7
-
作者声明不存在利益冲突
-
参考文献
(16) [1] LiuHF,ZhangF,LineaweaverWC.History and advancement of burn treatments[J].Ann Plast Surg,2017,78(2 Suppl 1):S2-8.DOI: 10.1097/SAP.0000000000000896. [2] LiH,YaoZ,TanJ,et al.Epidemiology and outcome analysis of 6325 burn patients: a five-year retrospective study in a major burn center in Southwest China[J].Sci Rep,2017,7:46066.DOI: 10.1038/srep46066. [3] SharmaA,AnumanthanG,ReyesM,et al.Epigenetic modification prevents excessive wound healing and scar formation after glaucoma filtration surgery[J].Invest Ophthalmol Vis Sci,2016,57(7):3381-3389.DOI: 10.1167/iovs.15-18750. [4] MooreLD,LeT,FanG.DNA methylation and its basic function[J].Neuropsychopharmacology,2013,38(1):23-38.DOI: 10.1038/npp.2012.112. [5] HorvathS,RajK.DNA methylation-based biomarkers and the epigenetic clock theory of ageing[J].Nat Rev Genet,2018,19(6):371-384.DOI: 10.1038/s41576-018-0004-3. [6] unknownAuthor. A bioinformatics workshop in a box[J].Nature,2018,554(7690):134.DOI: 10.1038/d41586-018-01424-4. [7] AltmanR.Current progress in bioinformatics 2016[J].Brief Bioinform,2016,17(1):1.DOI: 10.1093/bib/bbv105. [8] PeiG,ChenL,ZhangW.WGCNA Application to proteomic and metabolomic data analysis[J].Methods Enzymol,2017,585:135-158.DOI: 10.1016/bs.mie.2016.09.016. [9] SchierleHP,ScholzD,LemperleG.Elevated levels of testosterone receptors in keloid tissue: an experimental investigation[J].Plast Reconstr Surg,1997,100(2):390-395; discussion 396.DOI: 10.1097/00006534-199708000-00017. [10] LiuW,WangDR,CaoYL.TGF-beta: a fibrotic factor in wound scarring and a potential target for anti-scarring gene therapy[J].Curr Gene Ther,2004,4(1):123-136.DOI: 10.2174/1566523044578004. [11] LianN,LiT.Growth factor pathways in hypertrophic scars: molecular pathogenesis and therapeutic implications[J].Biomed Pharmacother,2016,84:42-50.DOI: 10.1016/j.biopha.2016.09.010. [12] NguyenJK,AustinE,HuangA,et al.The IL-4/IL-13 axis in skin fibrosis and scarring: mechanistic concepts and therapeutic targets[J].Arch Dermatol Res,2020,312(2):81-92.DOI: 10.1007/s00403-019-01972-3. [13] WynnTA.Cellular and molecular mechanisms of fibrosis[J].J Pathol,2008,214(2):199-210.DOI: 10.1002/path.2277. [14] HallamMJ,PittE,ThomasA,et al.Low-dose insulin as an antiscarring therapy in breast surgery: a randomized controlled trial[J].Plast Reconstr Surg,2018,141(4):476e-485e.DOI: 10.1097/PRS.0000000000004199. [15] FrikJ,Merl-PhamJ,PlesnilaN,et al.Cross-talk between monocyte invasion and astrocyte proliferation regulates scarring in brain injury[J].EMBO Rep,2018,19(5):e45294.DOI: 10.15252/embr.201745294. [16] XieY,OstrikerAC,JinY,et al.LMO7 is a negative feedback regulator of transforming growth factor β signaling and fibrosis[J].Circulation,2019,139(5):679-693.DOI: 10.1161/CIRCULATION-AHA.118.034615. -
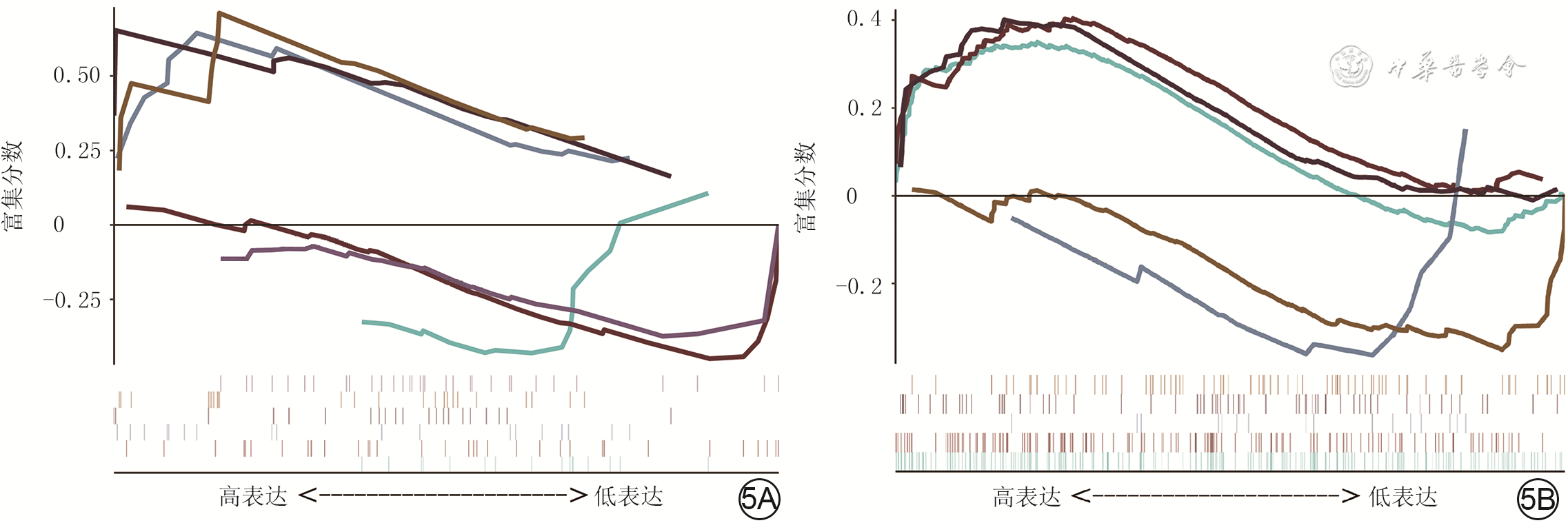
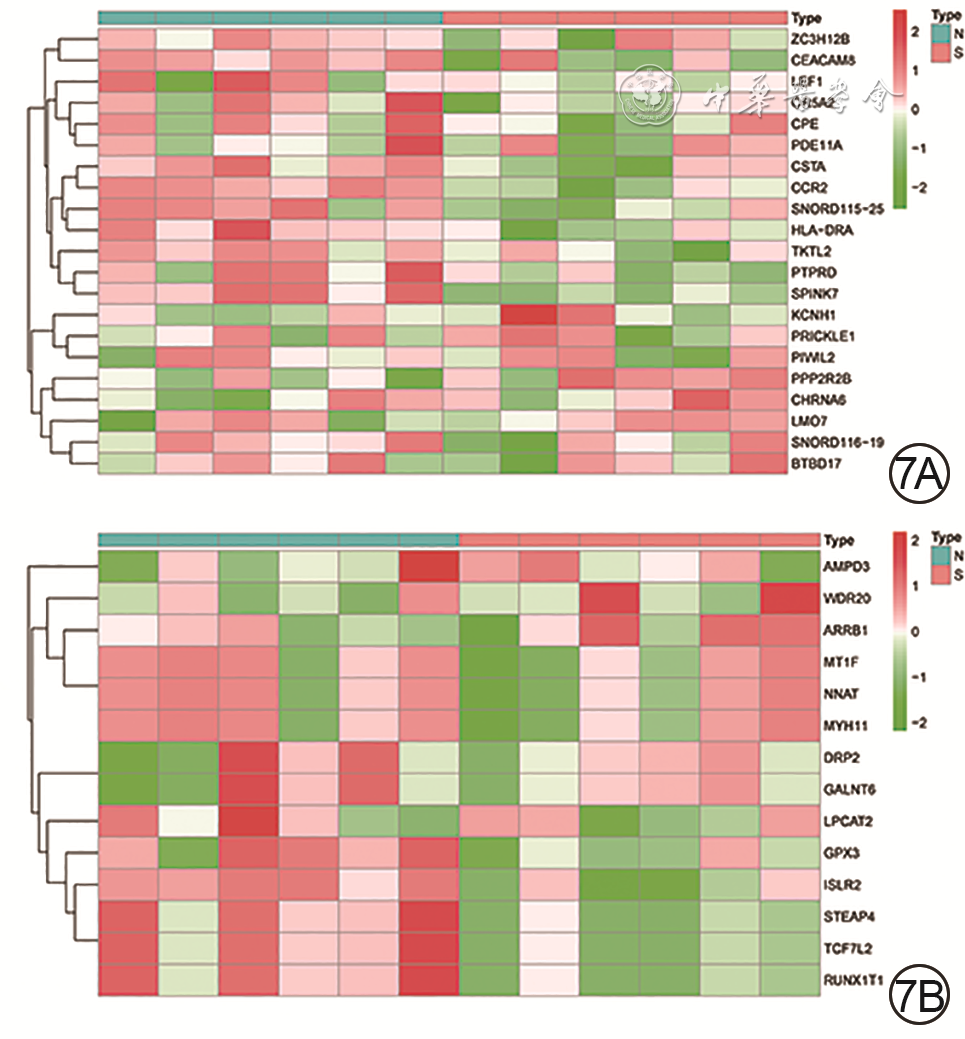
5 对烧伤后瘢痕组织相关性高的模块通过基因探针富集分析进行基因功能和通路分析。5A.基因本体论功能富集;5B.京都基因与基因组百科全书功能富集
注:图5A中的绿、棕、蓝、黑、浅棕、紫色曲线分别表示对核因子κB导入细胞核的负向调节、对白细胞介素1产生的正向调节、对α-氨基-3-羟基-5-甲基-4-异恶唑-丙酸酯选择性谷氨酸受体的活性调控、雄激素受体信号通路调控、脂肪酸氧化的调控、泛酸样蛋白结合酶的调节通路,图5B中的绿、棕、蓝、黑、浅棕色曲线分别表示细胞因子-细胞因子受体的相互作用、Janus激酶/信号转导与转录激活子信号通路、泛酸和辅酶A生物合成、过氧化物酶体增殖物激活受体信号途径、转化生长因子β信号途径
表1 烧伤后瘢痕组织相关数据集的详细信息
数据集 测序平台 Affymetrix eneChip文件 样本数(个) 瘢痕数(个) GSE137134 GPL13534-11288 1 6 6 GSE136906 GPL16686 2 6 6 GSE108110 GPL570 3 9 9 注:1、2、3分别为Illumina HumanMethylation450 BeadChip (HumanMethylation450_15017482)、[HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array [transcript (gene) version]、[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array 表2 烧伤后瘢痕组织与正常组织异常甲基化基因受试者操作特征曲线下面积及P值
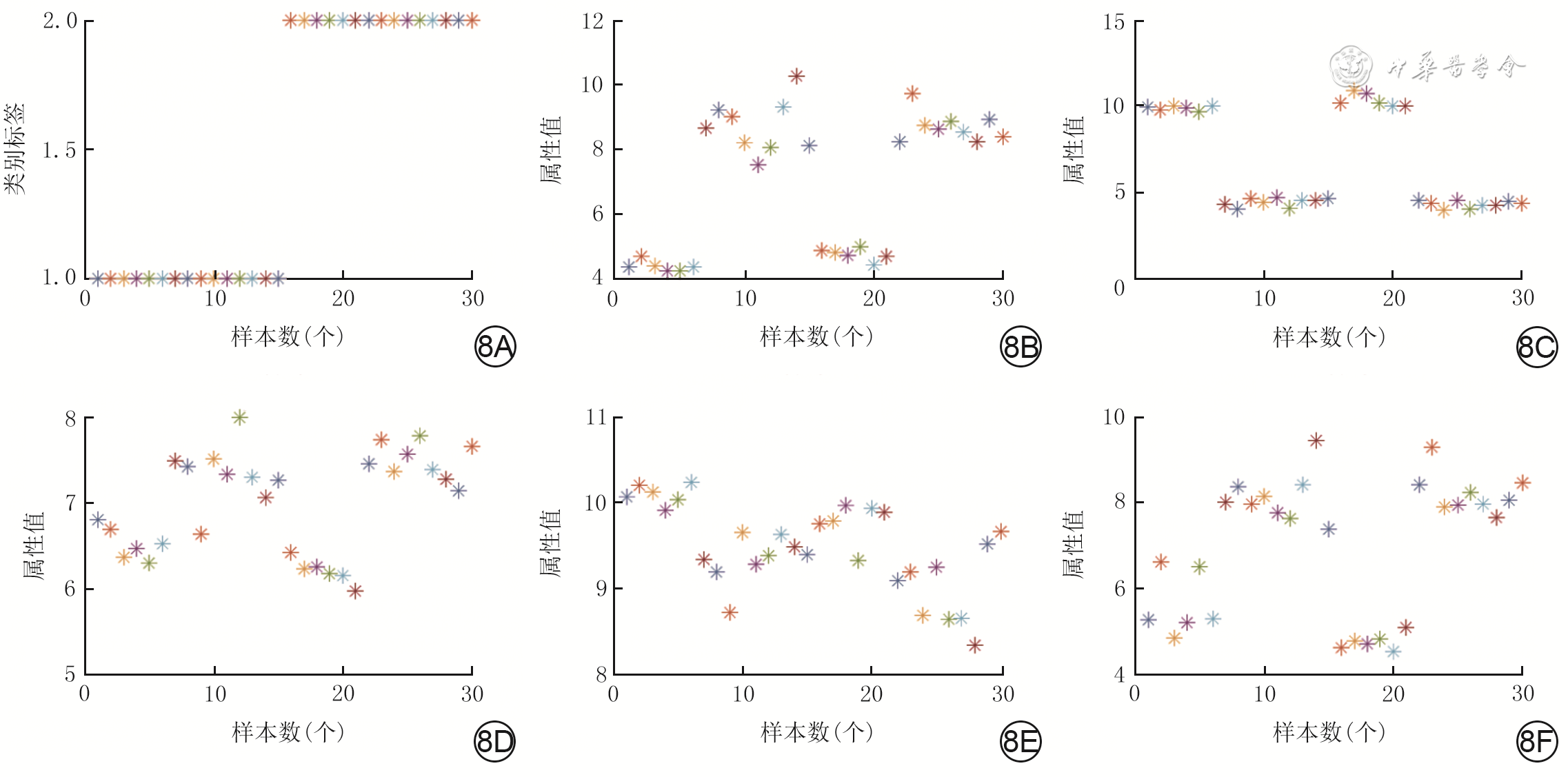
基因 曲线下面积 P值 CCR2 0.972 <0.001 LMO7 0.972 0.011 STEAP4 0.972 <0.001 NNAT 0.944 <0.001 TCF7L2 0.944 0.003 ZC3H12B 0.944 0.010 AMPD3 0.917 <0.001 BTBD17 0.917 0.039 PRICKLE1 0.917 <0.001 -







 下载:
下载: