Analysis of genomic information and biological characteristics of a bacteriophage against methicillin-resistant Staphylococcus aureus in patients with median sternal incision infection
-
摘要:
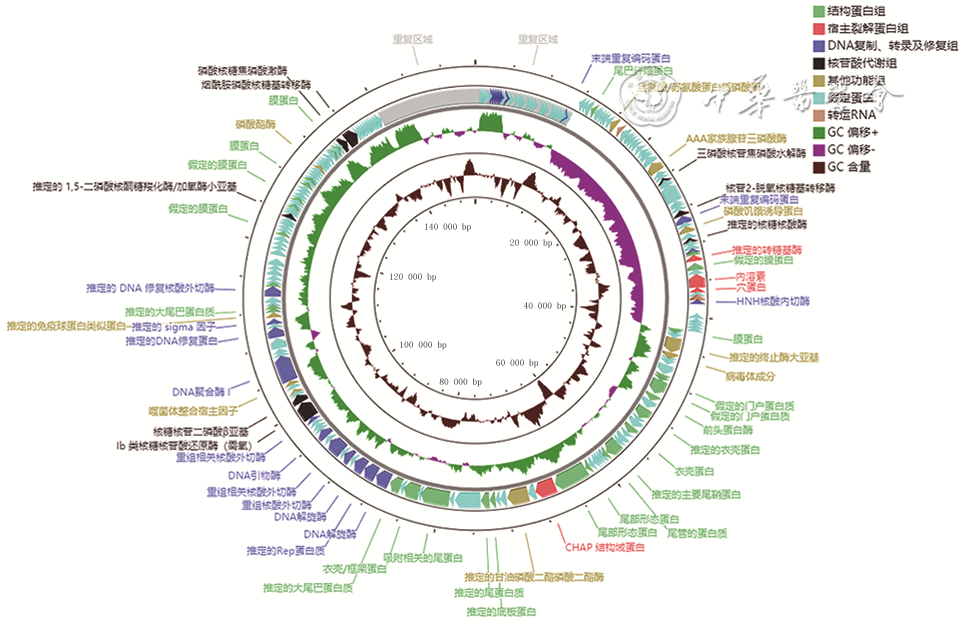
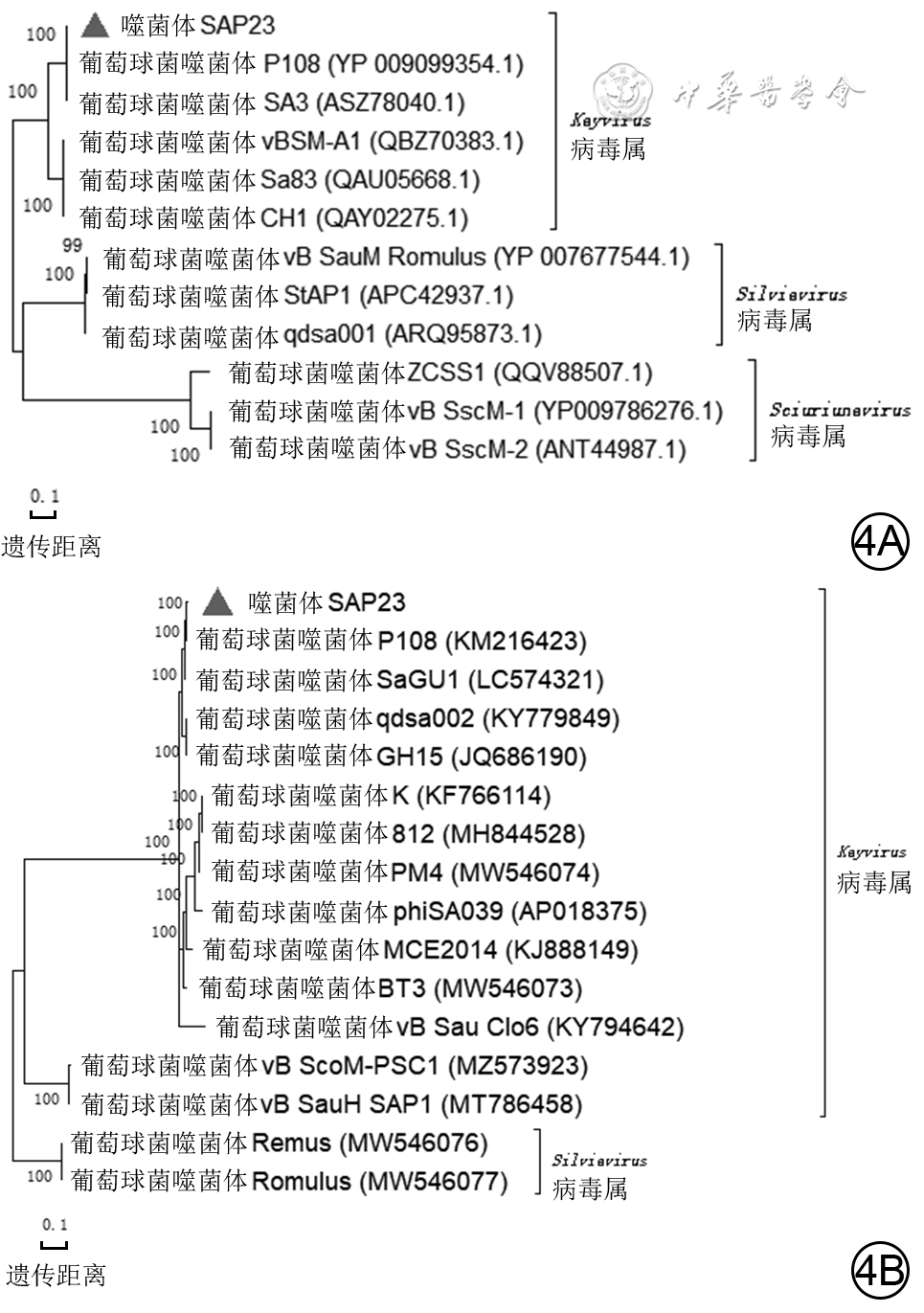
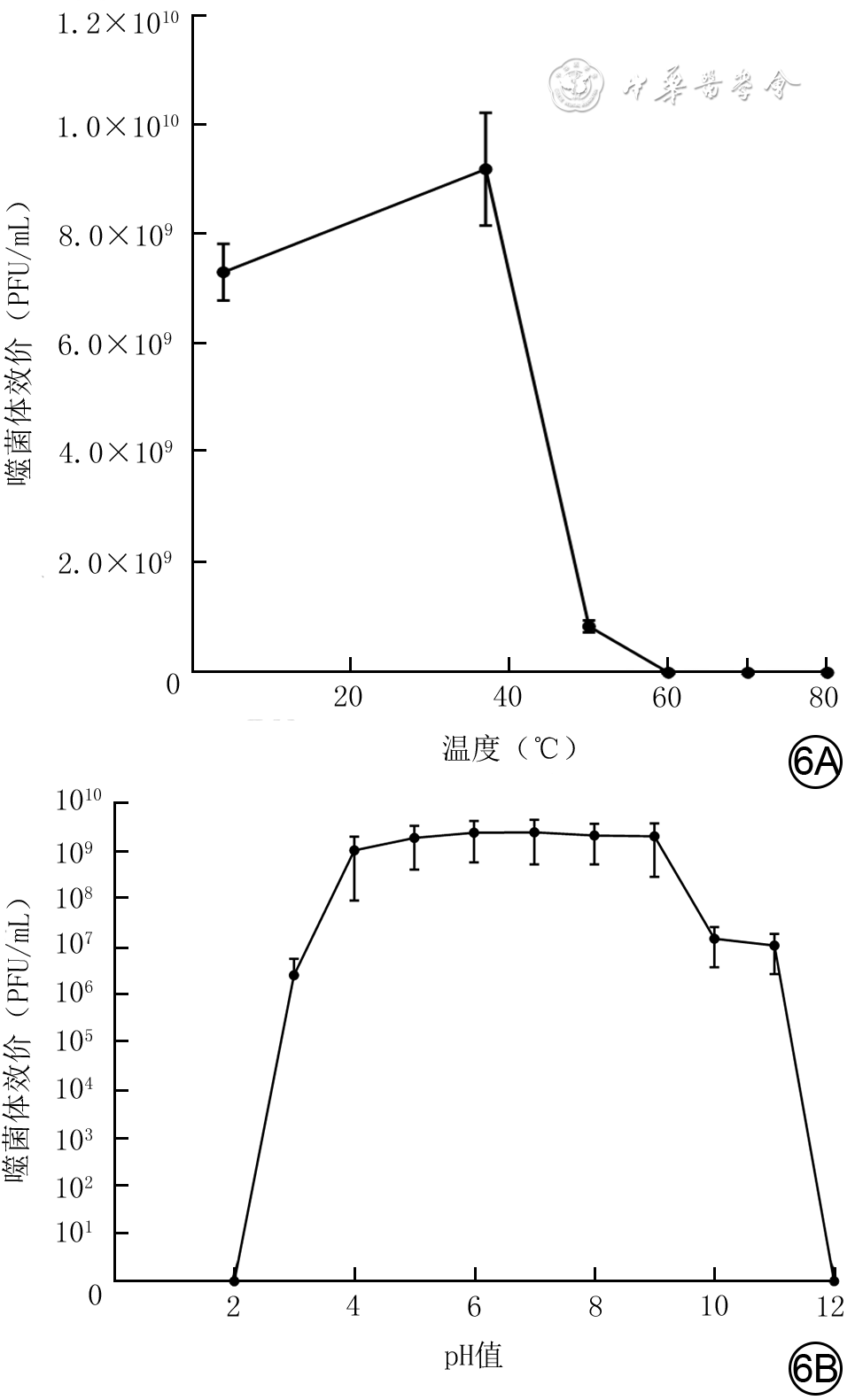
目的 分离提纯1株新型耐甲氧西林金黄色葡萄球菌(MRSA)的噬菌体,并对其基因组学信息和生物学特性进行分析。 方法 采用实验研究方法。取分离自陆军军医大学(第三军医大学)第二附属医院收治的1例胸骨正中切口感染的63岁女性患者创面的MRSA(下称宿主菌)液,采用污水共培养法和双层琼脂平板法从该院污水中分离提纯得到噬菌体,并命名为噬菌体SAP23,观察噬菌斑形态。采用磷钨酸负染法将噬菌体SAP23染色,采用透射电子显微镜观察其形态。采用十二烷基磺酸钠/蛋白酶裂解方案制备噬菌体SAP23 DNA,在Illumina NovaSeq PE150平台下进行全基因组测序,并完成序列组装、注释、系统发生树等基因组学分析。将噬菌体SAP23液分别按10.000 0、1.000 0、0.100 0、0.010 0、0.001 0、0.000 1感染复数与宿主菌液共培养4 h后,采用点滴法测定噬菌体效价,筛选最佳感染复数,此处及以下样本数均为3。按测得的最佳感染复数取噬菌体SAP23液与宿主菌液分别共同孵育5、10、15 min后,同前测定噬菌体效价,筛选最佳吸附时间。按测得的最佳感染复数取噬菌体SAP23液与宿主菌液按最佳吸附时间孵育后,分别于培养0(即刻)、5、10、15、20、30、40、50、60、80、100、120 min,同前测定噬菌体效价,绘制一步生长曲线。取噬菌体SAP23液分别在温度为4、37、50、60、70、80 ℃下,在pH值为2、3、4、5、6、7、8、9、10、11、12下孵育1 h,测定稳定性。取陆军军医大学(第三军医大学)微生物教研室储存的41株MRSA,完成噬菌体SAP23的宿主谱范围检测。 结果 噬菌体SAP23能在宿主菌双层琼脂板上形成透明噬菌斑。噬菌体SAP23头部是直径为(88±4)nm的多面体,其尾部长度为(279±21)nm、宽度为(22.6±2.6)nm。噬菌体SAP23基因组为全长151 618 bp的线状双链DNA,序列两端有11 681 bp的长末端重复序列,预测出220个开放阅读框,噬菌体可编码4个转运RNA,未预测出毒力因子或抗性基因,注释功能的噬菌体SAP23基因可分为5个组,GenBank登录号为MZ427930,噬菌体SAP23全基因组序列与共线性分析中的6个葡萄球菌噬菌体全基因组序列有5个局部共线区域,但在局部共线区域内部或外部存在差异。噬菌体SAP23属于Herelleviridae科Twortvirinae亚科Kayvirus病毒属。噬菌体SAP23的最佳感染复数为0.010 0,最佳吸附时间为10 min,潜伏期约为20 min,裂解期约为80 min;在4~37 ℃温度条件及pH值为4~9的条件中,稳定性较好。噬菌体SAP23可裂解41株MRSA中的3株。 结论 噬菌体SAP23为Herelleviridae科Twortvirinae亚科Kayvirus病毒属成员,潜伏期短,其对温度和酸碱耐受性好,可有效裂解MRSA,为不含毒力因子和抗性基因的新型烈性窄谱噬菌体。 -
关键词:
- 耐甲氧西林金黄色葡萄球菌 /
- 细菌噬菌体 /
- 基因组学 /
- 伤口感染 /
- 生物学特性
Abstract:Objective To isolate and purify a bacteriophage against methicillin-resistant Staphylococcus aureus (MRSA), and to analyze its genomic information and biological characteristics. Methods The experimental research methods were adopted. MRSA (hereinafter referred to as host bacteria) solution was collected from the wound of a 63-year-old female patient with the median sternum incision infection admitted to the Second Affiliated Hospital of Army Medical University (the Third Military Medical University). The bacteriophage, named bacteriophage SAP23 was isolated and purified from the sewage of the Hospital by sewage co-culture method and double-layer agar plate method, and the plaque morphology was observed. The morphology of bacteriophage SAP23 was observed by transmission electron microscope after phosphotungstic acid negative staining. The whole genome of bacteriophage SAP23 was sequenced with NovaSeq PE15 platform after its DNA was prepared by sodium dodecyl sulfonate/protease cleavage scheme, and genomic analysis including sequence assembly, annotation, and phylogenetic tree were completed. The bacteriophage SAP23 solution was co-incubated with the host bacterial solution for 4 h at the multiplicity of infection (MOI) of 10.000 0, 1.000 0, 0.100 0, 0.010 0, 0.001 0, and 0.000 1, respectively, and then the bacteriophage titer was measured by the drip plate method to select the optimal MOI, with here and the following sample numbers of 3. The bacteriophage SAP23 solution was co-incubated with the host bacterial solution at the optimal MOI for 5, 10, and 15 min, respectively, and the bacteriophage titer was measured by the same method as mentioned above to select the optimal adsorption time. After the bacteriophage SAP23 solution was co-incubated with the host bacterial solution at the optimal MOI for the optimal adsorption time, the bacteriophage titers were measured by the same method as mentioned above at 0 (immediately), 5, 10, 15, 20, 30, 40, 50, 60, 80, 100, and 120 min after culture, respectively, and a one-step growth curve was drawn. The bacteriophage SAP23 solution was incubated at 4, 37, 50, 60, 70, and 80 ℃ and pH 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, and 12 for 1 h, respectively, to determine its stability. A total of 41 MRSA strains stored in the Department of Microbiology of Army Medical University (the Third Military Medical University) were used to determine the host spectrum of bacteriophage SAP23. Results The bacteriophage SAP23 could form a transparent plaque on the host bacteria double-layer agar plate. The bacteriophage SAP23 has a polyhedral head with (88±4) nm in diameter and a tail with (279±21) nm in length and (22.6±2.6) nm in width. The bacteriophage SAP23 has a linear, double-stranded DNA with a full length of 151 618 bp and 11 681 bp long terminal repeats sequence in the sequence ends. There were 220 open reading frames predicted and the bacteriophage could encode 4 transfer RNAs, while no resistance genes or virulence factors were found. The annotation function of bacteriophage SAP23 genes could be divided into 5 groups. The GenBank accession number was MZ427930. According to the genomic collinearity analysis, there were 5 local collinear blocks in the whole genome between the bacteriophage SAP23 and the chosen 6 Staphylococcus bacteriophages, while within or outside the local collinear region, there were still some differences. The bacteriophage SAP23 belonged to the Herelleviridae family, Twortvirinae subfamily, and Kayvirus genus. The optimal MOI of bacteriophage SAP23 was 0.010 0, and the optimal adsorption time was 10 min. The bacteriophage SAP23 had a latent period of 20 min, and a growth phase of 80 min. The bacteriophage SAP23 was able to remain stable at the temperature between 4 and 37 ℃ and at the pH values between 4 and 9. The bacteriophage SAP23 could lyse 3 of the 41 tested MRSA strains. Conclusions The bacteriophage SAP23 is a member of the Herelleviridae family, Twortvirinae subfamily, and Kayvirus genus. The bacteriophage SAP23 has a good tolerance for temperature and acid-base and a short latent period, and can lyse MRSA effectively. The bacteriophage SAP23 is a new type of potent narrow-spectrum bacteriophage without virulence factors and resistance genes. -
(1)针对成年住院烧伤患者压力性损伤预防管理这一主题,通过系统检索文献并进行质量评价,提取、汇总了最佳证据并进行证据分级。
(2)该证据总结易于理解和传播,为医护人员提供了成年住院烧伤患者压力性损伤预防的实践依据。
Highlights:
(1)Aiming at the topic of prevention and management of pressure injury in adult hospitalized burn patients, this research extracted, summarized, and graded the best evidence through systematical search and quality evaluation of literature.
(2)The summary of evidence is easy to understand and disseminate, and provides a practical basis for health care personnel to prevent pressure injury in adult hospitalized burn patients.
根据世界卫生组织的数据,每年有180 000例与烧伤有关的患者死亡 [ 1] 。人体皮肤不仅是抵御外界环境的保护性屏障,还能防止体液、电解质和蛋白质的流失,并在钙代谢、维生素D3生成、体温调节和免疫系统中发挥重要作用 [ 2, 3] 。烧伤患者因皮肤屏障功能受损、长时间卧床、营养不良、严重水肿等原因,住院期间发生压力性损伤的风险较其他住院患者高 [ 4, 5] 。虽然目前指导压力性损伤预防的指南较多 [ 6, 7, 8] ,但适用人群较为广泛,尚缺乏针对烧伤患者压力性损伤预防的指南和共识。采取有效的预防措施能避免绝大多数压力性损伤事件的发生 [ 9, 10] 。因此,本研究采用文献计量学方法,针对“成年住院烧伤患者压力性损伤预防管理”这一主题,通过系统检索文献并评价,提取相关证据进行分级汇总,构建基于最佳证据的压力性损伤预防管理方案,以期帮助医护人员快速做出科学决策。
1. 资料与方法
1.1 问题的确立
基于如何预防成年住院烧伤患者压力性损伤的发生、如何提高成年住院烧伤患者压力性损伤的照护质量等问题,依照PIPOST模式将临床问题转化为循证问题 [ 11] ,其中证据应用的目标人群(P):成年住院烧伤患者,干预方法(I):压力性损伤的预防、干预,专业人员(P):烧伤科的护理人员,结局指标(O):烧伤患者压力性损伤的发生率、预防措施落实率、护理质量,证据应用场所(S):烧伤普通病房、烧伤ICU,证据类型(T):临床决策、证据总结、指南、系统评价、专家共识。
1.2 检索策略
按照循证检索资源的“6S”模型,依次检索临床决策系统、指南网站、专业学术组织网站及数据库。以“pressure injury/pressure ulcer/pressure ulcers/bedsore/bedsores/pressure sore/pressure sores/decubitus ulcers/decubitus ulcer/decubital ulcer、burn/burns/burning/burnt/burned”为英文检索词检索UpToDate、BMJ Best Practice临床决策系统,国际烧伤协会(ISBI)、美国烧伤协会、日本皮肤病协会等相关学术组织官方网站,美国压力性损伤专家咨询小组和欧洲压力性损伤专家咨询小组与环太平洋压力性损伤联盟合作的国际指南网站,MedSci、Joanna Briggs Institute(JBI) Evidence-Based Practice Database、Cochrane Library、Web of Science、Embase、PubMed等外文数据库。以“烧伤/压力性创伤/压力性损伤/压疮/压力性溃疡/褥疮”为检索词检索中国生物医学文献服务系统、中国知网、万方数据库、医脉通指南网等中文数据库,检索日期截至2023年2月21日。
1.3 文献入选标准
纳入标准:(1)研究对象为年龄≥18岁的烧伤患者;(2)研究内容涉及住院烧伤患者压力性损伤的预防和管理策略;(3)研究类型为临床决策、证据总结、指南、系统评价、专家共识;(4)发表语种为中文、英文。
排除标准:(1)重复发表的文献;(2)无法获取原文的文献;(3)已经更新过的旧版指南文献;(4)质量评价低的文献。
1.4 文献质量评价
由2名经过JBI循证护理中心系统培训的研究者根据相应的文献质量评价标准进行独立评价。当2人意见不一致时则与第3名研究者共同讨论,直至达成一致意见。
采用质量评价工具CASE(critical appraisal for summaries of evidence)进行证据总结的质量评价,共10个条目,均对每个条目进行“是/不完全是/否”的判定 [ 12] 。
采用2017年更新的《临床实践指南研究与评价系统Ⅱ》进行指南的质量评价,结果以6个领域标准化百分比表示,所有领域标准化百分比>60%为A级(推荐),≥3个领域标准化百分比<30%为C级(不推荐),其余情况为B级(修改完善后推荐) [ 13] ,最终纳入A级指南。
采用JBI循证卫生保健中心对应的标准进行专家共识的质量评价,共6个条目,均对每个条目进行“是/否/不清楚”的判定 [ 14] 。
1.5 证据提取和分级
由3名具备循证经验的烧伤和创面修复领域的研究者独立对最终纳入文献进行内容提取,参照文献[ 15]对纳入证据追溯的原始文献进行等级划分,证据级别共分为5级(1级最高,5级最低)。组织在烧伤领域、伤口护理领域工作10年以上、职称为中级及以上且熟悉本研究主题的1名烧伤领域医疗专家、2名烧伤领域护理专家、2名伤口护理专家、2名经JBI循证护理中心系统培训的护理专家共同组成专家组,进行证据级别推荐(A级推荐即强推荐,B级推荐即弱推荐)。
2. 结果
2.1 文献筛选结果
初步检索获得文献3 185篇,剔除重复文献后获得2 980篇,通过阅读文题和摘要后保留103篇,阅读全文复筛后最终纳入文献10篇,包括证据总结6篇 [ 16, 17, 18, 19, 20, 21] 、指南3篇 [ 6, 7, 8] 、专家共识1篇 [ 22] 。纳入文献的基本特征见 表1。
表1 纳入的10篇成年住院烧伤患者压力性损伤预防策略相关文献的基本特征表1. Basic characteristics of the included 10 papers about prevention strategies for pressure injury in adult hospitalized burn patients第1作者 发表年份 类型 主题 来源 Abdulsalam A [ 16] 2021 证据总结 烧伤患者压力性损伤的患病率 Joanna Briggs Institute Munn Z [ 17] 2021 证据总结 烧伤患者压力性损伤缓解策略 Joanna Briggs Institute Abdulsalam A [ 18] 2021 证据总结 烧伤患者压力性损伤预防(使用减压床垫) Joanna Briggs Institute Overall B [ 19] 2022 证据总结 成年压力性损伤的风险评估 Joanna Briggs Institute Marin T [ 20] 2022 证据总结 成年压力性损伤患者的营养支持 Joanna Briggs Institute Abdulsalam A [ 21] 2021 证据总结 手术患者压力性损伤的预防策略 Joanna Briggs Institute ISBI Practice Guidelines Committee [ 6] 2018 指南 烧伤患者的护理 ISBI官网 Cartotto R [ 7] 2023 指南 危重烧伤患者的早期康复运动 PubMed Kottner J [ 8] 2019 指南 压力性损伤患者的预防和治疗 NPUAP和EPUAP与PPPIA合作的国际指南网站 中国老年医学学会烧创伤分会 [ 22] 2022 专家共识 烧伤患者俯卧位治疗的管理 万方数据库 注:ISBI为国际烧伤协会,NPUAP为美国压力性损伤专家咨询小组,EPUAP为欧洲压力性损伤专家咨询小组,PPPIA为环太平洋压力性损伤联盟 2.2 文献质量评价结果
2.2.1 证据总结的质量评价结果
6篇证据总结 [ 16, 17, 18, 19, 20, 21] 评价为“是”的条目均≥7/10,见 表2,均予以纳入。
表2 纳入的6篇成年住院烧伤患者压力性损伤预防策略相关证据总结的质量评价结果表2. The quality evaluation results of the included 6 evidence summaries about prevention strategies of pressure injury in adult hospitalized burn patients第1作者 条目1 条目2 条目3 条目4 条目5 条目6 条目7 条目8 条目9 条目10 Abdulsalam A [ 16] 是 是 否 是 是 是 否 是 否 是 Munn Z [ 17] 是 是 否 是 是 是 是 是 否 是 Abdulsalam A [ 18] 是 是 否 是 是 是 是 是 否 是 Overall B [ 19] 是 是 否 是 是 是 是 是 否 是 Marin T [ 20] 是 是 否 是 是 是 是 是 否 是 Abdulsalam A [ 21] 是 是 否 是 是 是 是 是 否 是 注:条目1为“证据总结应用的范围和对象是否具体?”,条目2为“证据总结作者的身份是否清晰透明?”,条目3为“评审是否清晰透明?”,条目4为“证据分级是否清晰?”,条目5为“推荐意见是否清晰?”,条目6为“推荐意见引注是否恰当?”,条目7为“推荐意见是否有时效性?”,条目8为“证据总结条目是否适用于本研究人群评价?”,条目9为“文献检索是否全面?”,条目10为“是否有利益冲突声明?” 2.2.2 指南的质量评价结果
3篇指南 [ 6, 7, 8] 所有领域标准化百分比均>60%,质量评价为A级,见 表3,均予以纳入。
表3 纳入的3篇成年住院烧伤患者压力性损伤预防策略相关指南的质量评价各领域标准化百分比(%)表3. The quality evaluation results of the included 3 guidelines about prevention strategies of pressure injury in adult hospitalized burn patients2.2.3 专家共识的质量评价结果
1篇专家共识 [ 22] 明确标注了观点来源,观点来源于该领域有影响力的专家,所提出的观点以研究的人群利益为中心,陈述的结论是基于分析的结果且观点的表达具有逻辑性,参考了现有的其他文献,但未说明提出的观点与以往文献有不一致的地方,即专家共识质量评价标准的前5个条目评价均为“是”,第6个条目评价为“否”,整体质量较高,予以纳入。
2.3 最佳证据汇总与分级
2.3.1 压力性损伤预防的培训和监管
共3条证据,具体如下。(1)评估烧伤科医护人员关于压力性损伤预防的知识、态度、行为,针对烧伤科医护人员进行压力性损伤预防知识和技能培训 [ 19] (等级4,A级推荐)。(2)组织医院管理人员、医护人员、患者及其照护者参与压力性损伤预防计划的实施和监督 [ 8] (等级5,A级推荐)。(3)通过开展烧伤患者压力性损伤管理质量改进项目,降低压力性损伤的发生率 [ 8] (等级5,A级推荐)。
2.3.2 风险评估
共8条证据,具体如下。(1)建议在成年烧伤患者入院时,采用有效的量表,包括Norton压力性损伤风险评估表、Braden压力性损伤风险评估表、Waterlow压力性损伤风险评估表进行压力性损伤风险筛查 [ 19, 20] (等级3,A级推荐)。(2)建议在烧伤患者病情发生显著变化时,重新应用有效量表进行压力性损伤风险评分 [ 19] (等级3,A级推荐)。(3)建议在烧伤创面换药期间,进行压力性损伤风险评估 [ 19] (等级4,A级推荐)。(4)注意区分烧伤患者的压力性损伤和其他类型创面 [ 8] (等级5,A级推荐)。(5)烧伤患者压力性损伤危险因素包括老年、烧伤程度重、合并骨折或吸入性损伤、感染、多次手术等 [ 19] (等级3,A级推荐)。(6)建议重点评估烧伤患者的枕部、骶骨和脚后跟皮肤发生压力性损伤的风险 [ 16] (等级2,A级推荐)。(7)建议每天定时检查烧伤患者使用胃管、气管插管、夹板、弹力绷带处皮肤的颜色变化和水肿情况,及时调整上述医疗器具的固定位置和松紧度 [ 19] (等级4,A级推荐)。(8)建议在患者入院72 h内采用经过验证的工具筛查患者的营养状况,从而评估患者发生压力性损伤的风险 [ 19] (等级4,A级推荐)。
2.3.3 预防措施
共16条证据,具体如下。(1)推荐使用压力再分布床垫预防烧伤患者压力性损伤的发生 [ 19] (等级2,B级推荐)。(2)对于未合并严重呼吸系统损伤的成年烧伤患者,采用翻身床治疗有助于预防压力性损伤 [ 22] (等级5,A级推荐)。(3)针对烧伤患者制订并实施个体化的体位变更计划,翻身频率应考虑患者活动水平、活动能力、独立翻身的能力、皮肤耐受性、治疗状况、舒适度和疼痛情况 [ 20] (等级1,A级推荐)。(4)烧伤患者使用气垫床的情况下,每4小时翻身1次;对使用常规床垫卧床的压力性损伤高风险患者,至少每2小时翻身1次,被认为易受压力影响的烧伤患者应该更频繁地翻身 [ 19] (等级1,B级推荐)。(5)烧伤患者优先行30°侧卧,一般情况下不行90°侧卧 [ 20] (等级5,A级推荐)。(6)双足功能位摆放期间采取措施,如使用足跟保护装置缓解脚后跟压力 [ 19] (等级4,A级推荐)。(7)使用专用腿部抬高装置预防下肢皮肤移植部位和供皮部位发生压力性损伤 [ 19] (等级3,B级推荐)。(8)大便失禁患者肛周潮湿区域使用液体成膜剂或敷料等建立皮肤屏障,保持局部清洁干燥,预防失禁性皮炎 [ 19] (等级4,A级推荐)。(9)烧伤患者避免使用一次性尿布 [ 19] (等级4,A级推荐)。(10)鼓励烧伤患者多饮水,必要时通过静脉补充液体 [ 20] (等级5,A级推荐)。(11)烧伤患者受压皮肤干燥,可考虑外用保湿霜 [ 17] (等级4,B级推荐)。(12)对于手术中发生压力性损伤风险较大的烧伤患者,建议手术前对骨突出处预防性使用敷料 [ 21] (等级5,A级推荐)。(13)对于手术中发生压力性损伤风险较大的烧伤患者,术前应在手术台上覆盖1层减压垫,如高规格反应性泡沫床垫,或压力可交替变化的床垫,并记录使用效果 [ 21] (等级5,A级推荐)。(14)建议制订并实施烧伤患者早期活动方案,预防压力性损伤的发生 [ 6, 7] (等级2,B级推荐)。(15)建议针对烧伤患者实施个体化的营养方案,确保患者有足够的营养摄入量 [ 19] ,合理补充维生素和矿物质,预防压力性损伤的发生 [ 19, 20] (等级1,A级推荐)。(16)对烧伤患者及其照护者进行健康教育,内容包括压力性损伤发生原因和危险因素、预防措施、自我护理方法,并提供社会心理支持指导 [ 8, 22] (等级5,A级推荐)。
3. 讨论
3.1 证据总结的形成过程科学严谨
本研究通过PIPOST [ 11] 界定研究问题,聚焦“成年住院烧伤患者压力性损伤预防管理”进行文献检索,文献质量由2名经过JBI循证护理中心系统培训的研究者进行评价,确保证据来源的科学性。证据提取阶段,由3名具备循证经验的烧伤和创面修复领域的研究者独立对最终纳入文献进行内容提取,从压力性损伤预防的培训与监管、风险评估、预防措施3个方面分类汇总27条证据并注明每条证据的等级和推荐强度,为临床医护人员开展烧伤患者压力性损伤预防管理提供了科学依据。
3.2 加强医护人员培训和压力性损伤预防工作质量管理
住院患者压力性损伤发生率是评价护理工作质量的敏感指标之一 [ 23, 24] 。加强医院组织层面管理是降低压力性损伤发生率的前提和保障 [ 24] 。本文总结了3条相关证据,强调规范化培训体系和质量管理对烧伤患者压力性损伤的预防具有重要意义 [ 8, 19] 。2条证据建议对烧伤科医护人员进行压力性损伤相关知识的培训,组织医院管理人员、医护人员、患者及其照护者参与压力性损伤预防计划的实施和质量管理,形成闭环管理 [ 8, 19] 。证据应用过程中,建议使用医院电子病历系统来追踪压力性损伤风险评估和预防措施的落实率,促进压力性损伤预防工作质量提升。
3.3 规范和动态开展烧伤患者压力性损伤风险评估
风险评估是预防压力性损伤发生的第1步 [ 8] 。Braden压力性损伤风险评估表、Norton压力性损伤风险评估表和Waterlow压力性损伤风险评估表各有评估的侧重点。虽然多项研究表明Braden压力性损伤风险评估表是评估烧创伤患者压力性损伤发生风险的重要工具 [ 9, 10] ,但也有1项单中心回顾性研究证实Braden量表对烧伤患者压力性损伤风险辨别能力有限,需要进一步探讨并建立适用于住院烧伤患者的压力性损伤风险评估量表 [ 9] 。现有证据建议结合烧伤患者病情特点和发生压力性损伤的高风险因素开展全面评估。由于烧伤患者表皮完整性受到损害,长时间的体位固定以及敷料覆盖,会导致创面受压且不易观察 [ 8] 。在烧伤创面换药期间,应进行压力性损伤风险评估,注意鉴别压力性损伤和其他类型创面 [ 8] 。烧伤患者治疗期间管道固定位置不当、使用不当或应用后未规范监测,均会导致局部组织受损,进一步发展为医疗器械相关性压力性损伤(medical device related pressure injury,MDRPI),推荐开展MDRPI的评估 [ 19] 。烧伤后,机体处于较强应激状态,分解代谢过程加快,容易导致患者营养不良 [ 2] ,建议常规开展烧伤患者营养风险评估,为实施科学的营养管理和预防压力性损伤提供依据 [ 19] 。
3.4 采取有效措施预防烧伤患者压力性损伤的发生
烧伤患者压力性损伤预防措施包括合理选择床具、及时变更体位、皮肤管理、术中压力性损伤预防管理、落实营养支持和早期康复活动等。减压床垫通过减少烧伤患者身体和接触面之间的压力,并将压力重新分配到面积大的承重表面上,预防压力性损伤的发生,但目前尚无高质量证据表明哪种床垫能兼顾最佳的临床结局和成本效益,因此研究小组讨论后给予B级推荐。翻身床在烧伤科临床中应用普遍,在未合并严重呼吸系统损伤的前提下,烧伤患者行翻身床治疗有助于预防压力性损伤 [ 22] 。医用悬浮床可提供干爽和碱性环境减少局部受压 [ 23] ,但此次未收集到相关的高质量证据,提示可进一步收集相关原始研究证据。在体位选择方面,30°侧卧体位是减少身体受到的摩擦力和剪切力的有效措施,建议优先采用 [ 20] 。建议烧伤患者应采取比每2小时1次更为频繁的翻身 [ 19] ,但由于重症烧伤患者多合并严重并发症,建议临床医护人员充分考虑患者病情严重程度、已采取的预防措施及患者耐受度等因素,确定个体化的体位变换频率,保障患者安全。通过容量管理和外用保湿霜,以保持皮肤适当的湿润度并预防烧伤患者大小便失禁,避免使用一次性尿布、及时清理排泄物、会阴及肛周皮肤使用液体成膜剂或敷料建立皮肤屏障,预防失禁性皮炎 [ 17, 19, 20] 。重度烧伤患者需经历清创、皮片移植、皮瓣转移等多次手术,对于手术中发生压力性损伤风险较大的烧伤患者,根据手术时长、术中体位,合理使用保护性敷料、手术台上使用减压设备预防术中压力性损伤 [ 21] 。证据推荐通过早期活动预防烧伤患者发生压力性损伤 [ 6, 7] ,但ISBI指南同时也提出,尚缺乏足够的原始研究证明在ICU环境中启动早期活动能够减少成年危重烧伤患者压力性损伤的发生 [ 6] ,经研究小组讨论,给予“B级推荐”。
3.5 本研究存在的不足与未来研究方向
本研究通过循证方法学汇总了成年住院烧伤患者压力性损伤预防策略的最佳证据,包括压力性损伤预防的团队培训和组织管理、风险评估、防范措施。未来,拟将这些证据应用到临床,开展循证实践,制订适合我国成年住院烧伤患者的压力性损伤预防策略的最佳方案,降低成年住院烧伤患者压力性损伤的发生率。
张建:酝酿和设计实验、实施研究、采集数据、分析/解释数据、文章起草;燕荣帅:酝酿和设计实验、对文章的知识性内容作批评性审阅、指导;杨子晨:酝酿和设计实验、对文章的知识性内容作批评性审阅、获取研究经费、指导、支持性贡献;石茜:实施研究、采集数据;李翔、毛彤春:酝酿和设计实验、采集数据;张一鸣:酝酿和设计实验、文章起草、对文章的知识性内容作批评性审阅、行政、技术或材料支持、指导、支持性贡献所有作者均声明不存在利益冲突 -
参考文献
(63) [1] WangPH, HuangBS, HorngHC, et al.Wound healing[J].J Chin Med Assoc,2018, 81(2):94-101. DOI: 10.1016/j.jcma.2017.11.002. [2] WilkinsonHN, HardmanMJ.Wound healing: cellular mechanisms and pathological outcomes[J].Open Biol,2020, 10(9):200223. DOI: 10.1098/rsob.200223. [3] SarhanWA, AzzazyHME, El-SherbinyIM.Honey/chitosan nanofiber wound dressing enriched with allium sativum and cleome droserifolia: enhanced antimicrobial and wound healing activity[J].ACS Appl Mater Interfaces,2016, 8(10):6379-6390. DOI: 10.1021/acsami.6b00739. [4] HanG, CeilleyR.Chronic wound healing: a review of current management and treatments[J].Adv Ther,2017, 34(3):599-610. DOI: 10.1007/s12325-017-0478-y. [5] SiddiquiAR,BernsteinJM.Chronic wound infection: facts and controversies[J].Clin Dermatol,2010,28(5):519-526.DOI: 10.1016/j.clindermatol.2010.03.009. [6] LakhundiS, ZhangKY. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology[J].Clin Microbiol Rev,2018,31(4):e0020-18.DOI: 10.1128/CMR.00020-18. [7] SongRh,YuB,FriedrichD,et al.Naphthoquinone-derivative as a synthetic compound to overcome the antibiotic resistance of methicillin-resistant S. aureus[J].Commun Biol,2020,3(1):529.DOI: 10.1038/s42003-020-01261-0. [8] Hernández-AristizábalI, Ocampo-IbáñezID.Antimicrobial peptides with antibacterial activity against vancomycin-resistant Staphylococcus aureus strains: classification, structures, and mechanisms of action[J].Int J Mol Sci,2021, 22(15):7927. DOI: 10.3390/ijms22157927. [9] CrockerTF, BrownL, LamN, et al. Information provision for stroke survivors and their carers[J].Cochrane Database Syst Rev,2021,11(11):CD001919.DOI: 10.1002/14651858.CD001919.pub4. [10] CisekAA, DąbrowskaI, GregorczykKP, et al.Phage therapy in bacterial infections treatment: one hundred years after the discovery of bacteriophages[J].Curr Microbiol,2017, 74(2):277-283. DOI: 10.1007/s00284-016-1166-x. [11] KortrightKE, ChanBK, KoffJL, et al.Phage therapy: a renewed approach to combat antibiotic-resistant bacteria[J].Cell Host Microbe,2019, 25(2):219-232. DOI: 10.1016/j.chom.2019.01.014. [12] SummersWC.The strange history of phage therapy[J].Bacteriophage,2012, 2(2):130-133. DOI: 10.4161/bact.20757. [13] SarkerSA,SultanaS,ReutelerG,et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh[J].EBioMedicine,2016,4:124-137.DOI: 10.1016/j.ebiom.2015.12.023. [14] FurfaroLL, PayneMS, ChangBJ.Bacteriophage therapy: clinical trials and regulatory hurdles[J].Front Cell Infect Microbiol,2018, 8:376. DOI: 10.3389/fcimb.2018.00376. [15] JaultP,LeclercT,JennesS,et al.Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial[J].Lancet Infect Dis,2019,19(1):35-45.DOI: 10.1016/S1473-3099(18)30482-1. [16] LeitnerL, UjmajuridzeA, ChanishviliN, et al.Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: a randomised, placebo-controlled, double-blind clinical trial[J].Lancet Infect Dis,2021,21(3):427-436.DOI: 10.1016/S1473-3099(20)30330-3. [17] CarvalhoC, CostaAR, SilvaF, et al.Bacteriophages and their derivatives for the treatment and control of food-producing animal infections[J].Crit Rev Microbiol,2017,43(5):583-601.DOI: 10.1080/1040841X.2016.1271309. [18] MichelsonD, GrundmanM, MagnusonK, et al.Randomized, placebo controlled trial of NPT088, a phage-derived, amyloid-targeted treatment for Alzheimer's disease[J].J Prev Alzheimers Dis,2019, 6(4):228-231. DOI: 10.14283/jpad.2019.37. [19] YangZC, LiuXZ, ShiYL, et al.Characterization and genome annotation of a newly detected bacteriophage infecting multidrug-resistant Acinetobacter baumannii[J].Arch Virol,2019,164(6):1527-1533.DOI: 10.1007/s00705-019-04213-0. [20] LuSG, LeS,TanYL, et al.Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages[J].PLoS One,2013,8(5):e62933.DOI: 10.1371/journal.pone.0062933. [21] GarneauJR, DepardieuF, FortierLC, et al.PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data[J].Sci Rep,2017,7(1):8292.DOI: 10.1038/s41598-017-07910-5. [22] BesemerJ,LomsadzeA,BorodovskyM.GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions[J].Nucleic Acids Res,2001,29(12):2607-2618.DOI: 10.1093/nar/29.12.2607. [23] BrettinT,DavisJJ,DiszT,et al.RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes[J].Sci Rep,2015,5:8365.DOI: 10.1038/srep08365. [24] AzizRK,BartelsD,BestAA,et al.The RAST Server: rapid annotations using subsystems technology[J].BMC Genomics,2008,9:75.DOI: 10.1186/1471-2164-9-75. [25] ArndtD,GrantJR,MarcuA,et al. PHASTER: a better, faster version of the PHAST phage search tool[J].Nucleic Acids Res,2016,44(W1):W16-21.DOI: 10.1093/nar/gkw387. [26] LoweTM, EddySR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence[J].Nucleic Acids Res,1997,25(5):955-964.DOI: 10.1093/nar/25.5.955. [27] LagesenK,HallinP,RødlandEA,et al.RNAmmer: consistent and rapid annotation of ribosomal RNA genes[J].Nucleic Acids Res,2007,35(9):3100-3108.DOI: 10.1093/nar/gkm160. [28] DarlingACE, MauB, BlattnerFR,et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements[J].Genome Res,2004,14(7):1394-1403.DOI: 10.1101/gr.2289704. [29] AnisimovaM.Evolutionary genomics: statistical and computational methods[M]. New York: Springer New York,2019: 121-147. [30] El-ArabiTF,GriffithsMW,SheYM,et al.Genome sequence and analysis of a broad-host range lytic bacteriophage that infects the Bacillus cereus group[J].Virol J,2013,10:48.DOI: 10.1186/1743-422X-10-48. [31] 吴丽飞,王兆飞,王中华,等.高效裂解多重耐药金黄色葡萄球菌的噬菌体分离及裂解酶的制备[J].中国动物传染病学报,2021,29(3):1-9. DOI: 10.19958/j.cnki.cn31-2031/s.2021.03.001. [32] YangS,YangY,CuiSX,et al.Chitosan-polyvinyl alcohol nanoscale liquid film-forming system facilitates MRSA-infected wound healing by enhancing antibacterial and antibiofilm properties[J].Int J Nanomedicine,2018,13:4987-5002.DOI: 10.2147/IJN.S161680. [33] DouJL, JiangYW, XieJQ, et al.New is old, and old is new: recent advances in antibiotic-based, antibiotic-free and ethnomedical treatments against methicillin-resistant Staphylococcus aureus wound infections[J].Int J Mol Sci,2016, 17(5):617. DOI: 10.3390/ijms17050617. [34] MaciejewskaB,OlszakT,Drulis-KawaZ.Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: an ambitious and also a realistic application?[J].Appl Microbiol Biotechnol,2018,102(6):2563-2581.DOI: 10.1007/s00253-018-8811-1. [35] MorozovaVV, VlassovVV, TikunovaNV.Applications of bacteriophages in the treatment of localized infections in humans[J].Front Microbiol,2018, 9:1696. DOI: 10.3389/fmicb.2018.01696. [36] OoiML, DrillingAJ, MoralesS, et al.Safety and tolerability of bacteriophage therapy for chronic rhinosinusitis due to Staphylococcus aureus[J].JAMA Otolaryngol Head Neck Surg,2019, 145(8):723-729. DOI: 10.1001/jamaoto.2019.1191. [37] FishR, KutterE, BryanD, et al.Resolving digital staphylococcal osteomyelitis using bacteriophage-a case report[J].Antibiotics (Basel),2018, 7(4):87. DOI: 10.3390/antibiotics7040087. [38] 胡福泉.噬菌体的过去、现在与未来[J].西南医科大学学报,2021,44(5):417-424.DOI: 10.3969/j.issn.2096-3351.2021.05.001. [39] RegeimbalJM, JacobsAC, CoreyBW, et al.Personalized therapeutic cocktail of wild environmental phages rescues mice from Acinetobacter baumannii wound infections[J].Antimicrob Agents Chemother,2016, 60(10):5806-5816. DOI: 10.1128/aac.02877-15. [40] GillJJ, HymanP.Phage choice, isolation, and preparation for phage therapy[J].Curr Pharm Biotechnol,2010, 11(1):2-14. DOI: 10.2174/138920110790725311. [41] KrupovicM,DutilhBE,AdriaenssensEM,et al.Taxonomy of prokaryotic viruses: update from the ICTV bacterial and archaeal viruses subcommittee[J].Arch Virol,2016,161(4):1095-1099.DOI: 10.1007/s00705-015-2728-0. [42] ŁobockaM,HejnowiczMS,DąbrowskiK,et al.Genomics of staphylococcal Twort-like phages--potential therapeutics of the post-antibiotic era[J].Adv Virus Res,2012,83:143-216.DOI: 10.1016/B978-0-12-394438-2.00005-0. [43] AzamAH, TanjiY.Peculiarities of Staphylococcus aureus phages and their possible application in phage therapy[J].Appl Microbiol Biotechnol,2019, 103(11):4279-4289. DOI: 10.1007/s00253-019-09810-2. [44] Głowacka-RutkowskaA, UlatowskaM, EmpelJ, et al.A Kayvirus distant homolog of staphylococcal virulence determinants and VISA biomarker is a phage lytic enzyme[J].Viruses,2020, 12(3):292. DOI: 10.3390/v12030292. [45] DonovanDM,LardeoM,Foster-FreyJ.Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin[J].FEMS Microbiol Lett,2006,265(1):133-139.DOI: 10.1111/j.1574-6968.2006.00483.x. [46] PaulVD,RajagopalanSS,SundarrajanS,et al.A novel bacteriophage tail-associated muralytic enzyme (TAME) from Phage K and its development into a potent antistaphylococcal protein[J].BMC Microbiol,2011,11:226.DOI: 10.1186/1471-2180-11-226. [47] GuJM,XuW,LeiLC,et al.LysGH15, a novel bacteriophage lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection[J].J Clin Microbiol,2011,49(1):111-117.DOI: 10.1128/JCM.01144-10. [48] KaurJ, SinghP, SharmaD, et al. A potent enzybiotic against methicillin-resistant Staphylococcus aureus[J].Virus Genes,2020, 56(4):480-497. DOI: 10.1007/s11262-020-01762-4. [49] CahillJ, YoungR.Phage lysis: multiple genes for multiple barriers[J].Adv Virus Res,2019, 103:33-70. DOI: 10.1016/bs.aivir.2018.09.003. [50] LindenSB,ZhangH,HeselpothRD,et al.Biochemical and biophysical characterization of PlyGRCS, a bacteriophage endolysin active against methicillin-resistant Staphylococcus aureus[J].Appl Microbiol Biotechnol,2015,99(2):741-752.DOI: 10.1007/s00253-014-5930-1. [51] 高明明,刘慧莹,李璞媛,等.金黄色葡萄球菌噬菌体vB_SauH_IME522的分离鉴定及全基因组分析[J].第三军医大学学报,2020,42(3):229-240.DOI: 10.16016/j.1000-5404.201909016. [52] ShimamoriY, PramonoAK, KitaoT, et al. Isolation and characterization of a novel phage SaGU1 that infects Staphylococcus aureus clinical isolates from patients with atopic dermatitis[J].Curr Microbiol,2021, 78(4):1267-1276. DOI: 10.1007/s00284-021-02395-y. [53] Bailly-BechetM, VergassolaM, RochaE. Causes for the intriguing presence of tRNAs in phages[J].Genome Res,2007,17(10):1486-1495.DOI: 10.1101/gr.6649807. [54] NunesA, RibeiroDR, MarquesM,et al.Emerging roles of tRNAs in RNA virus infections[J].Trends Biochem Sci,2020,45(9):794-805.DOI: 10.1016/j.tibs.2020.05.007. [55] McCallinS, SarkerSA, BarrettoC,et al.Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects[J].Virology,2013,443(2):187-196.DOI: 10.1016/j.virol.2013.05.022. [56] QuirósP,Colomer-LluchM,Martínez-CastilloA,et al.Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples[J].Antimicrob Agents Chemother,2014,58(1):606-609.DOI: 10.1128/AAC.01684-13. [57] 靳晓东,张聪慧,钟江.两株新的金黄色葡萄球菌烈性噬菌体的生物学特性和基因组学研究[J].微生物与感染,2018,13(6):335-341.DOI: 10.3969/j.issn.1673-6184.2018.06.003. [58] JiJW, LiuQ, WangR, et al. Identification of a novel phage targeting methicillin-resistant Staphylococcus aureus in vitro and in vivo[J].Microbial Pathogenesis,2020, 149:104317. DOI: https://doi.org/10.1016/j.micpath.2020.104317. [59] GutiérrezD, VandenheuvelD, MartínezB, et al.Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono- and dual-species Staphylococcal biofilms[J].Appl Environ Microbiol,2015, 81(10):3336-3348. DOI: 10.1128/aem.03560-14. [60] FengTT, LeptihnS, DongK, et al. JD419, a Staphylococcus aureus phage with a unique morphology and broad host range[J].Front Microbiol,2021, 12:602902. DOI: 10.3389/fmicb.2021.602902. [61] DoubJB, NgVY, JohnsonAJ, et al.Salvage bacteriophage therapy for a chronic MRSA prosthetic joint infection[J].Antibiotics (Basel),2020, 9(5):241. DOI: 10.3390/antibiotics9050241. [62] JikiaD,ChkhaidzeN,ImedashviliE,et al.The use of a novel biodegradable preparation capable of the sustained release of bacteriophages and ciprofloxacin, in the complex treatment of multidrug-resistant Staphylococcus aureus-infected local radiation injuries caused by exposure to Sr90[J].Clin Exp Dermatol,2005,30(1):23-26.DOI: 10.1111/j.1365-2230.2004.01600.x. [63] LuongT, SalabarriaAC, RoachDR.Phage therapy in the resistance era: where do we stand and where are we going?[J].Clin Ther,2020, 42(9):1659-1680. DOI: 10.1016/j.clinthera.2020.07.014. -
表1 胸骨正中切口感染患者耐甲氧西林金黄色葡萄球菌噬菌体SAP23对41株耐甲氧西林金黄色葡萄球菌裂解情况
菌株编号 噬菌斑 菌株编号 噬菌斑 菌株编号 噬菌斑 菌株编号 噬菌斑 DP100 - CY6 ++ NF99 - G15 - CY15 - NF7 - SY14 - G26 - CY14 - NF70 - SY19 - G25 - CY19 - NF71 - SY33 - G31 - CY18 - NF73 - SY17 ++ G34 - CY5 - NF75 - SY5 - G13 - CY11 - NF8 - SY13 - G21 - CY16 - NF84 - SY23 - G17 - CY7 - NF88 - SY6 - G36 - CY8 - NF90 + SY15 - G16 - CY20 - 注:“++”表示可见透明噬菌斑,“+”表示可见混浊噬菌斑,“-”表示未见噬菌斑 -







 下载:
下载:


 下载:
下载: