Investigation on the growth factor regulatory network of dermal fibroblasts in mouse full-thickness skin defect wounds based on single-cell RNA sequencing
-
摘要:
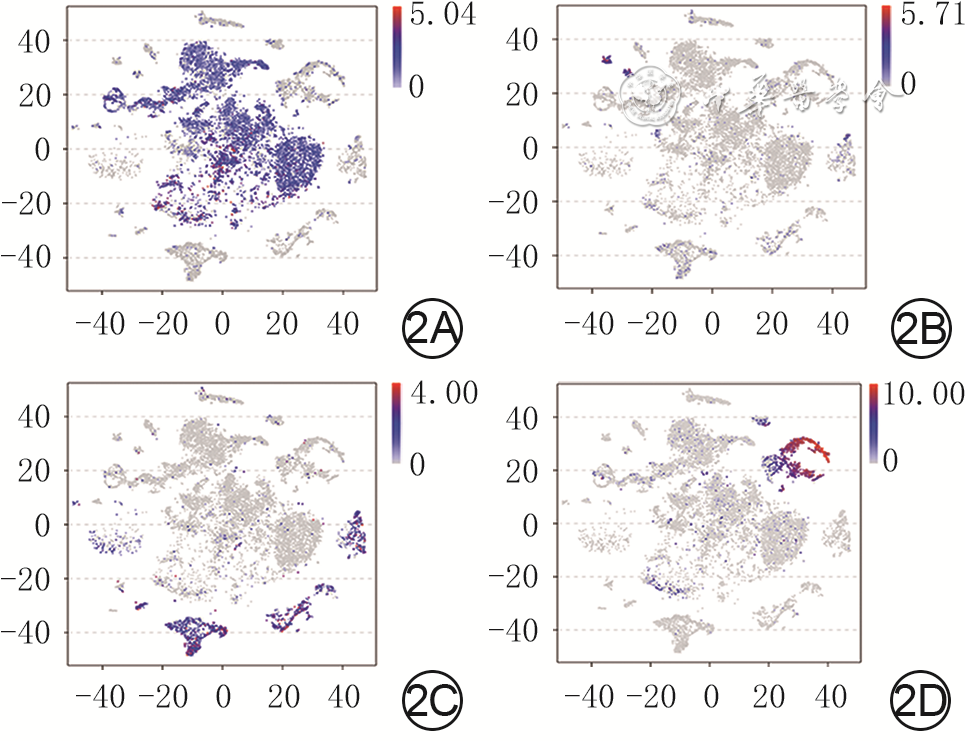
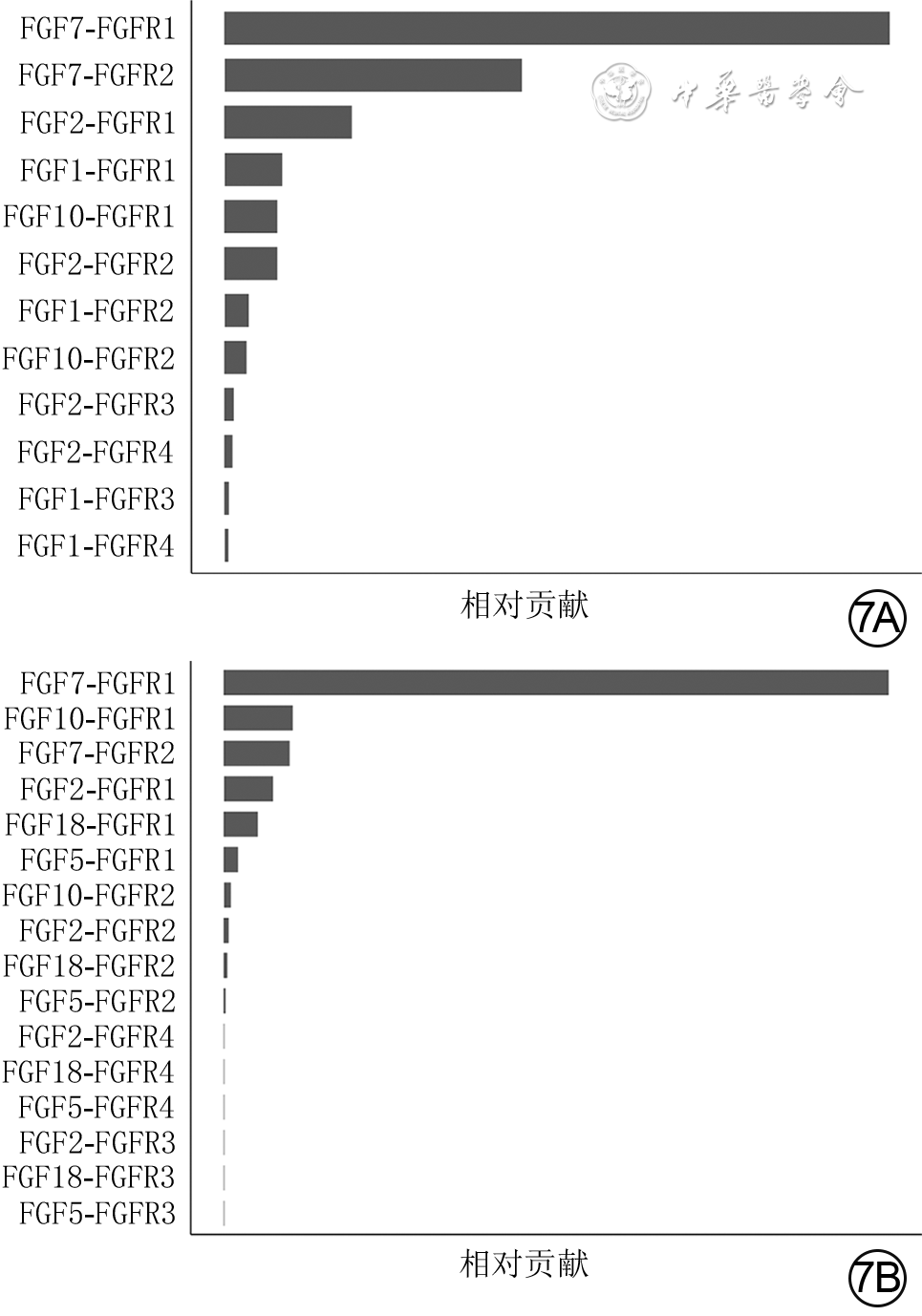
目的 基于单细胞RNA测序探讨小鼠全层皮肤缺损创面中真皮成纤维细胞(dFb)的异质性与生长因子调控网络。 方法 采用实验研究方法。取5只8周龄雄性健康C57BL/6小鼠(鼠龄、性别、品系下同)正常皮肤组织,另取5只背部全层皮肤缺损小鼠伤后7 d创面组织,用胶原酶D和DNA酶Ⅰ消化组织获得细胞悬液,用10x Genomics平台构建测序文库,用Illumina Novaseq6000测序仪进行单细胞RNA测序。采用R4.1.1软件的Seurat 3.0程序分析获得2种组织细胞的基因表达矩阵,采用按细胞群、细胞来源、标记皮肤中主要细胞基因分类的二维tSNE云图进行可视化展示。根据已有文献报道和CellMarker数据库检索情况,分析2种组织细胞的基因表达矩阵中标志基因表达情况,对各细胞群进行编号和定义。将基因表达矩阵和细胞分群信息导入R4.1.1软件的CellChat 1.1.3程序,分析2种组织中细胞间通信以及创面组织血管内皮生长因子(VEGF)、血小板衍生生长因子(PDGF)、表皮生长因子(EGF)、成纤维细胞生长因子(FGF)信号通路中的细胞间通信,2种组织中FGF的各亚型与FGF受体(FGFR)各亚型的两两配对(以下简称FGF配受体对)对FGF信号网络的相对贡献,2种组织中相对贡献排名前2的FGF配受体对信号通路中的细胞间通信。取1只健康小鼠正常皮肤组织和1只全层皮肤缺损小鼠伤后7 d创面组织,行多重免疫荧光染色,检测FGF7蛋白的表达与分布及其与二肽基肽酶4(DPP4)、干细胞抗原1(SCA1)、平滑肌肌动蛋白(SMA)和PDGF受体α(PDGFRα)的蛋白共定位表达。 结果 健康小鼠正常皮肤组织和全层皮肤缺损小鼠伤后7 d创面组织中均包含25个细胞群,但2种组织中各细胞群中细胞数不一致。PDGFRα、血小板内皮细胞黏附分子1、淋巴管内皮透明质酸受体1、受体型蛋白酪氨酸磷酸酶C、角蛋白10和角蛋白79基因在二维tSNE云图上均有各自明确的分布,分别指示特定的细胞群。将25个细胞群按C0~C24编号,分为9个dFb亚群和16个非dFb群,dFb亚群包括C0间质祖细胞、C5脂肪前体细胞、C13具有收缩能力的肌肉细胞相关成纤维细胞等,非dFb群包括C3中性粒细胞、C8 T细胞、C18红细胞等。与健康小鼠正常皮肤组织比较,全层皮肤缺损小鼠伤后7 d创面组织中细胞间通信更多更密集,其中细胞间通信强度排前3位的细胞群为dFb亚群C0、C1、C2,这3个dFb亚群与创面组织中其他细胞群之间均有通信。在全层皮肤缺损小鼠伤后7 d创面组织中,VEGF信号主要由dFb亚群C0发送、脉管相关细胞群C19和C21接收,PDGF信号主要由周细胞C14发送、多个dFb亚群接收,EGF信号主要由角质形成细胞亚群C9和C11发送、dFb亚群C0接收,FGF信号的主要发送者和接收者均为dFb亚群C6。健康小鼠正常皮肤组织和全层皮肤缺损小鼠伤后7 d创面组织FGF信号网络中的FGF配受体对相对贡献,均是FGF7-FGFR1排在首位,排名第2的分别是FGF7-FGFR2、FGF10-FGFR1;与正常皮肤组织比较,创面组织FGF7-FGFR1信号通路中的细胞间通信更多,而FGF7-FGFR2和FGF10-FGFR1信号通路中的细胞间通信稍有减少或无明显变化;创面组织FGF7-FGFR1信号通路中的细胞间通信强于FGF7-FGFR2、FGF10-FGFR1信号通路;在2种组织中,FGF7信号均主要由dFb亚群C0与C1和C2发送、dFb亚群C6和C7接收。与健康小鼠正常皮肤组织比较,全层皮肤缺损小鼠伤后7 d创面组织中FGF7蛋白表达更多;在正常皮肤组织中,FGF7蛋白主要表达于皮肤间质层且在靠近真皮白色脂肪组织附近也有表达;在2种组织中,FGF7蛋白与DPP4、SCA1蛋白共定位表达于皮肤间质层中,与PDGFRα蛋白共定位表达于dFb中,与SMA蛋白无共定位表达,其中创面组织中的共定位表达多于正常皮肤组织。 结论 小鼠全层皮肤缺损创面愈合过程中的dFb存在高异质性,为多种生长因子潜在的主要分泌或接收细胞群落,与生长因子信号通路之间存在紧密、复杂的联系;FGF7-FGFR1信号通路是创面愈合过程中的主要FGF信号通路,靶向调控多个dFb亚群。 Abstract:Objective To explore the heterogeneity and growth factor regulatory network of dermal fibroblasts (dFbs) in mouse full-thickness skin defect wounds based on single-cell RNA sequencing. Methods The experimental research methods were adopted. The normal skin tissue from 5 healthy 8-week-old male C57BL/6 mice (the same mouse age, sex, and strain below) was harvested, and the wound tissue of another 5 mice with full-thickness skin defect on the back was harvested on post injury day (PID) 7. The cell suspension was obtained by digesting the tissue with collagenase D and DNase Ⅰ, sequencing library was constructed using 10x Genomics platform, and single-cell RNA sequencing was performed by Illumina Novaseq6000 sequencer. The gene expression matrices of cells in the two kinds of tissue were obtained by analysis of Seurat 3.0 program of software R4.1.1, and two-dimensional tSNE plots classified by cell group, cell source, and gene labeling of major cells in skin were used for visual display. According to the existing literature and the CellMarker database searching, the expression of marker genes in the gene expression matrices of cells in the two kinds of tissue was analyzed, and each cell group was numbered and defined. The gene expression matrices and cell clustering information were introduced into CellChat 1.1.3 program of software R4.1.1 to analyze the intercellular communication in the two kinds of tissue and the intercellular communication involving vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), epidermal growth factor (EGF), and fibroblast growth factor (FGF) signal pathways in the wound tissue, the relative contribution of each pair of FGF subtypes and FGF receptor (FGFR) subtypes (hereinafter referred to as FGF ligand receptor pairs) to FGF signal network in the two kinds of tissue, and the intercellular communication in the signal pathway of FGF ligand receptor pairs with the top 2 relative contributions in the two kinds of tissue. The normal skin tissue from one healthy mouse was harvested, and the wound tissue of one mouse with full-thickness skin defect on the back was harvested on PID 7. The multiple immunofluorescence staining was performed to detect the expression and distribution of FGF7 protein and its co-localized expression with dipeptidyl peptidase 4 (DPP4), stem cell antigen 1 (SCA1), smooth muscle actin (SMA), and PDGF receptor α (PDGFRα) protein. Results Both the normal skin tissue of healthy mice and the wound tissue of full-thickness skin defected mice on PID 7 contained 25 cell groups, but the numbers of cells in each cell group between the two kinds of tissue were different. Genes PDGFRα, platelet endothelial cell adhesion molecule 1, lymphatic endothelial hyaluronic acid receptor 1, receptor protein tyrosine phosphatase C, keratin 10, and keratin 79 all had distinct distributions on two-dimensional tSNE plots, indicating specific cell groups respectively. The 25 cell groups were numbered by C0-C24 and divided into 9 dFb subgroups and 16 non-dFb groups. dFb subgroups included C0 as interstitial progenitor cells, C5 as adipose precursor cells, and C13 as contractile muscle cells related fibroblasts, etc. Non-dFb group included C3 as neutrophils, C8 as T cells, and C18 as erythrocytes, etc. Compared with that of the normal skin tissue of healthy mice, the intercellular communication in the wound tissue of full-thickness skin defected mice on PID 7 was more and denser, and the top 3 cell groups in intercellular communication intensity were dFb subgroups C0, C1, and C2, of which all communicated with other cell groups in the wound tissue. In the wound tissue of full-thickness skin defected mice on PID 7, VEGF signals were mainly sent by the dFb subgroup C0 and received by vascular related cell groups C19 and C21, PDGF signals were mainly sent by peripheral cells C14 and received by multiple dFb subgroups, EGF signals were mainly sent by keratinocyte subgroups C9 and C11 and received by the dFb subgroup C0, and the main sender and receiver of FGF signals were the dFb subgroup C6. In the relative contribution rank of FGF ligand receptor pairs to FGF signal network in the normal skin tissue of healthy mice and the wound tissue of full-thickness skin defected mice on PID 7, FGF7-FGFR1 was the top 1, and FGF7-FGFR2 or FGF10-FGFR1 was in the second place, respectively; compared with those in the normal skin tissue, there was more intercellular communication in FGF7-FGFR1 signal pathway, while the intercellular communication in FGF7-FGFR2 and FGF10-FGFR1 signal pathways decreased slightly or did not change significantly in the wound tissue; the intercellular communication in FGF7-FGFR1 signal pathway in the wound tissue was stronger than that in FGF7-FGFR2 or FGF10-FGFR1 signal pathway; in the two kinds of tissue, FGF7 signal was mainly sent by dFb subgroups C0, C1, and C2, and received by dFb subgroups C6 and C7. Compared with that in the normal skin tissue of healthy mouse, the expression of FGF7 protein was higher in the wound tissue of full-thickness skin defected mouse on PID 7; in the normal skin tissue, FGF7 protein was mainly expressed in the skin interstitium and also expressed in the white adipose tissue near the dermis layer; in the two kinds of tissue, FGF7 protein was co-localized with DPP4 and SCA1 proteins and expressed in the skin interstitium, co-localized with PDGFRα protein and expressed in dFbs, but was not co-localized with SMA protein, with more co-localized expression of FGF7 in the wound tissue than that in the normal skin tissue. Conclusions In the process of wound healing of mouse full-thickness skin defect wound, dFbs are highly heterogeneous, act as potential major secretory or receiving cell populations of a variety of growth factors, and have a close and complex relationship with the growth factor signal pathways. FGF7-FGFR1 signal pathway is the main FGF signal pathway in the process of wound healing, which targets and regulates multiple dFb subgroups. -
Key words:
- Wound healing /
- Skin /
- Dermis /
- Fibroblasts /
- Fibroblast growth factors /
- Single-cell RNA sequencing /
- Growth factor /
- Intercellular communication
-
(1)足底内侧游离皮瓣整复手掌电烧伤创面及瘢痕挛缩符合整复外科“组织近似原则”,在同源无毛皮肤修复的同时提供更好的手部抓握力,相比其他皮瓣拥有更好的耐用性和美观性。
(2)带皮神经的足底内侧游离皮瓣可以通过血流桥接的形式重建手掌远端血运和感觉神经。
(3)皮瓣供区隐蔽,且切取过程不损伤足底主干血管及神经,对皮瓣供区功能无明显影响。
Highlights:
(1)The medial plantar free flap for repairing electric burn wound and scar contracture in the palm was in line with the "like with like principle" of plastic surgery, and provided better grip strength of hand while repairing with homologous hairless skin, and had better durability and aesthetics than other flaps.
(2)The medial plantar free flap with cutaneous nerve could be used to reconstruct the blood transport and sensory nerve of the distal palm through the form of flow-through.
(3)The flap donor site was concealed, and the excision process did not damage the main blood vessels or nerves of the plantar, and had no obvious impact on the function of flap donor area.
手部电烧伤以手掌部为重,手掌部皮肤厚、电阻高,电烧伤常使手掌深部的软组织坏死甚至骨坏死,即便是手掌电烧伤后软组织缺损面积不大,经换药或皮片移植修复后仍会遗留严重的瘢痕挛缩畸形 [ 1] 。修复手掌皮肤软组织缺损的方法较多,但由于手掌具有皮肤结构复杂、感觉丰富、功能多样、大量垂直纤维束与掌腱膜相连导致皮肤滑动性小且修复要求高等特点,整复手掌电烧伤后创面及瘢痕挛缩一直是一项极具挑战的工作 [ 2] 。以往采用的游离皮片移植术往往会导致手掌耐磨性差,甚至出现严重的瘢痕挛缩畸形等并发症而饱受诟病 [ 3] 。随着显微外科技术的发展,传统的游离皮瓣能修复手掌创面,但存在术后外观臃肿、感觉及功能恢复差、供区损伤重等不足 [ 4] 。手掌皮肤与足底内侧皮肤组织结构类似,采用足底内侧游离皮瓣修复手掌皮肤软组织缺损是较为理想的方案 [ 5] 。为此,本研究团队采用吻合皮神经的足底内侧皮瓣游离移植整复手掌电烧伤创面及瘢痕挛缩,取得了良好的临床效果。
1. 对象与方法
本回顾性观察性研究符合《赫尔辛基宣言》的基本原则和要求。
1.1 入选标准
纳入标准:(1)电烧伤致手掌部创面或瘢痕挛缩,清创或瘢痕切除松解后伴腱性组织甚至骨组织外露,形成创面面积为3 cm×3 cm~8 cm×8 cm者;(2)采用吻合皮神经的足底内侧游离皮瓣修复者。排除标准:(1)术后随访时间不足3个月者;(2)术中资料、随访资料不全者。
1.2 临床资料
2020年1月—2023年1月,空军军医大学第一附属医院收治6例符合入选标准的高压电烧伤致手掌创面或瘢痕挛缩患者,其中电烧伤创面患者4例且均为男性,电烧伤后手掌瘢痕挛缩患者2例且男女各1例;年龄35~55岁(平均43岁);累及右手者4例、左手者2例;致伤电压为380 V~10 kV。4个电烧伤创面合并屈肌腱部分变性,2个瘢痕挛缩切除松解后创面伴有腱性组织外露。手术时间为入院后1~7 d。
1.3 治疗方法
1.3.1 皮瓣移植术前准备
术前常规完善血常规、凝血、肝肾功能等实验室检查。患者于气管插管全身麻醉下取仰卧位行手术,对4例患者的电烧伤创面行急诊清创探查修复术,去除失活组织,保留间生态组织、腱性组织及重要血管神经。2例瘢痕挛缩患者于入院后1~7 d接受瘢痕切除彻底松解修复术。清创及瘢痕切除后创面面积为5.0 cm×3.0 cm~8.0 cm×7.0 cm。
1.3.2 皮瓣设计与切取
在足底内侧非负重区标记内踝前线与足底内侧缘的交点,然后从这一交点出发标记一条直线到第1、2跖骨头之间,为足底内侧皮瓣的轴线。应用手持多普勒超声血流探测仪在此线的中点附近探查皮瓣穿支血管的穿出点并标记为皮瓣的轴心点。根据创面大小拓取皮瓣样布并将边缘线扩大0.5 cm后设计皮瓣大小。从皮瓣内侧缘切开皮肤并保留1根或2根优势浅静脉,然后向远端游离足够长度后结扎、切断备用。从足底腱膜表面掀起皮瓣达𧿹展肌,牵开𧿹展肌与趾短屈肌间隙寻找足底内侧动脉血管穿支及其伴行的足底内侧皮神经并予以保留 [ 6, 7] 。从皮瓣外侧缘切开后在足掌腱膜表面向内侧缘会师,切取时应注意保留皮瓣内侧部分垂直的皮系韧带与掌腱膜的连续性。游离足够长度穿支血管,保留足底内侧神经主干并无损伤束间分离足够长度的足底内侧皮神经。彻底游离皮瓣,仅保留足底内侧血管蒂,检查皮瓣血运良好后断蒂,本组患者皮瓣切取面积为5.5 cm×3.5 cm~8.5 cm×7.5 cm。皮瓣供区创面行腹部全厚皮移植修复后加压包扎,石膏固定踝关节于中立位。
1.3.3 皮瓣移植
修整皮瓣的多余脂肪组织使皮瓣厚度与行清创或瘢痕切除后的手掌创面相匹配,将皮瓣远端与手掌创面边缘固定,于显微镜下将足底内侧穿支动脉血管与患手桡动脉或尺动脉掌浅支血管行端端吻合,将穿支静脉血管与患手桡动脉或尺动脉伴行静脉行端端吻合,将足底内侧皮神经与正中神经或尺神经掌浅支近端外膜行端端吻合以重建皮瓣感觉。血管吻合完成后检查皮瓣血运恢复情况,待皮瓣充血反应正常后间断缝合创缘,于皮瓣下放置引流片3~5条。
1.3.4 术后处理及随访
术后常规给予抗炎、抗血管痉挛及抗血栓形成等对症支持治疗,保持患者病房室温>25 ℃,预留皮瓣观察窗并密切观察皮瓣血运变化。术后48 h拔除引流片。患者卧床1周。皮瓣供区皮片移植1周后换药,12 d拆线,踝关节石膏固定2周。下床后抬高患肢3周并穿戴弹力袜减轻皮瓣供区肢体远端水肿及瘢痕增生。出院后定期门诊随访。
1.4 观测指标
术后观察皮瓣和皮片存活情况。患者平均随访时间>12个月,随访观察皮瓣外形、质地及皮瓣供区情况,评估患手握持功能恢复情况。于末次随访时,测量皮瓣两点辨别觉距离并按照中华医学会手外科学会上肢部分功能评定试用标准 [ 8] 评定皮瓣感觉恢复情况,采用皮瓣术后功能评价量表评价皮瓣功能恢复情况,优:皮瓣成活好、外形匹配度高、感觉基本正常,良:皮瓣部分成活、感觉部分恢复,差:皮瓣未成活、感觉未恢复。
2. 结果
2.1 总体情况
术后5个皮瓣存活良好;1个皮瓣远端部分坏死,清创后移植大腿外侧中厚皮修复。皮瓣供区皮片均存活良好。患者均获随访,随访3~24个月,皮瓣不臃肿,质地、色泽好,与周围组织匹配度高;皮瓣供区无明显瘢痕挛缩发生;患手握持功能良好。末次随访时,皮瓣两点辨别觉距离为6~8 mm,皮瓣感觉恢复:5个皮瓣恢复到S3 +级、1个皮瓣恢复到S3级,皮瓣功能评定:优5个、良1个,患者基本恢复正常的生活和工作。
2.2 典型病例
例1
男,47岁,全身多处被10 kV高压电烧伤后3 h急诊入院。入院后行左手电烧伤创面清创探查修复术,清创后创面面积约7.5 cm×4.0 cm,伴屈肌腱变性。术中根据探查结果选择患手左侧尺动脉掌浅支为受区主要吻合血管,然后切取面积为8.0 cm×4.5 cm的同侧足底内侧游离皮瓣并移植于左手掌电烧伤创面,显微镜下将皮瓣足底内侧穿支动脉血管与患手尺动脉掌浅支血管行端端吻合,并将穿支静脉血管与患手尺动脉伴行静脉行端端吻合,将足底内侧皮神经与正中神经掌浅支近端外膜行端端吻合。移植腹部全厚皮修复皮瓣供区创面。行术后皮瓣和皮片均存活良好。术后随访,皮瓣外形、色泽好,皮瓣供区无明显瘢痕,患手握持功能好。术后6个月末次随访时,皮瓣两点辨别觉距离为7 mm,功能评定为优,感觉恢复到S3 +级。见 图1。
例2
男,37岁,左手掌380 V电烧伤行植皮术后瘢痕挛缩7个月入院。入院后2 d行左手掌瘢痕切除松解修复术,形成7.5 cm×4.5 cm大小创面,伴创基掌浅弓血管及腱性组织外露。切取面积为8.0 cm×5.0 cm的同侧足底内侧游离皮瓣,显微镜下将足底内侧穿支动脉血管与患手桡动脉掌浅支血管行端端吻合,将穿支静脉血管与患手桡动脉伴行静脉行端端吻合。将足底内侧皮神经与正中神经掌浅支近端外膜行端端吻合。移植腹部全厚皮修复皮瓣供区创面。术后皮瓣和皮片均存活良好。术后随访,皮瓣外形好,质地与周围组织匹配度高,皮瓣供区无明显瘢痕,患手握持功能好。术后10个月末次随访时,皮瓣两点辨别觉距离为8 mm,功能评定为优,感觉恢复到S3 +级。见 图2。
3. 讨论
在电烧伤意外发生时,由于上肢接触电流的机会大,手掌更是容易被电烧伤,造成明显的软组织损伤 [ 9] ,手掌因其功能重要性和组织结构特殊性在受伤后常常造成严重的后果 [ 10] 。手掌部位电烧伤创面往往深达肌肉深面甚至伴有腱性组织和骨质的破坏 [ 11] 。皮片移植并不适合手掌深度电烧伤创面的修复 [ 12] 。传统皮瓣如带蒂的腹部皮瓣 [ 13] 和前臂逆行岛状瓣 [ 14] 是常用的修复手部创面的皮瓣,但因其组织结构与手掌皮肤差异较大易出现诸多并发症,从而导致手术效果欠佳 [ 2] 。
在多数情况下,手掌皮肤软组织缺损修复绝非简单的皮瓣修复或单纯的感觉功能重建,手掌皮肤软组织缺损修复后的耐用性更加重要 [ 15] 。足底内侧皮肤在结构上与手掌最相近,有较厚的角化层及脂肪垫,垂直的皮系韧带将皮肤与掌腱膜相连,使足底内侧皮肤不易滑动,有利于其移植修复手掌皮肤软组织后恢复手部持、捏等功能,同时耐磨抗压 [ 16] 。足底皮肤的乳头层内有与手掌皮肤结构相似的感觉神经末梢及感受器,所以感觉十分灵敏。足底内侧区为非负重区,位置隐蔽,足底内侧皮瓣最早由被学者提出应用于足跟部皮肤软组织缺损的重建,该皮瓣切取不会损伤足部重要血管,且切取后对足部的血运及站立行走等功能并无明显影响 [ 17] 。显微外科技术的发展为足底内侧皮瓣游离移植奠定了一定的基础,而对足底及手掌解剖结构的进一步研究则为足底内侧游离皮瓣移植修复手掌创面提供了坚实的解剖学基础。足底内侧皮瓣也从起初的筋膜皮瓣发展到后续的感觉皮瓣,带有功能重建作用的肌皮瓣、穿支皮瓣、静脉皮瓣、折叠皮瓣以及和足内侧皮瓣联合的嵌合皮瓣等 [ 18] 。针对本组病例本研究团队采用吻合皮神经的足底内侧游离皮瓣整复手掌电烧伤创面及瘢痕挛缩,皮瓣移植后长期随访结果显示,皮瓣的外形、色泽良好,感觉恢复良好,同时患手握持功能恢复良好;皮瓣供区亦无明显瘢痕挛缩形成或影响肢体活动及行走的情况出现。
要推广应用本术式除了需要提高显微操作技术,还应该注意以下事项:(1)术前应采用手持多普勒超声血流探测仪探测足底内侧穿支血管的穿出点及走行,明确是否存在血管变异、闭塞或缺如等情况;(2)足底皮肤致密,皮瓣切取后回缩很少,因此在设计时皮瓣边缘仅较创面边缘扩大0.5 cm即可;(3)足底内侧皮神经与正中神经或尺神经掌浅支吻合和皮瓣的血管吻合同等重要,缺一不可;(4)皮瓣切取时应注意保留皮瓣内侧部分垂直的皮系韧带与掌腱膜的连续性,将垂直的皮系韧带与受区腱膜或创面基底缝合以更好地对抗皮瓣移植后的滑动;(5)皮瓣切取范围远端不宜超过第1跖骨头侧缘,近端不超过内踝尖,外缘不超过足外侧负重点内缘,以免损伤足部负重区,遗留功能障碍;(6)足底内侧神经是胫神经的重要分支,是足底感觉的主要神经,手术时应将足底内侧神经主干留于原位以保留前足感觉。
本术式优点如下:(1)足底内侧游离皮瓣整复手掌电烧伤创面及瘢痕挛缩符合整复外科“组织近似原则”,在同源无毛皮肤修复的同时提供更好的手部抓握力,相比其他皮瓣拥有更好的耐用性和美观性;(2)带皮神经的足底内侧游离皮瓣可以通过血流桥接的形式重建手掌远端血运和感觉神经;(3)皮瓣供区隐蔽,且位于足部非负重区域,对皮瓣供区功能无明显影响;(4)皮瓣切取不损伤主干血管与神经,且皮瓣回流静脉粗大,吻合难度相对较低。然而,本术式同样存在一些无法回避的不足:(1)足底内侧皮瓣可切取的范围有限,仅限于足弓非负重区;(2)穿支血管蒂长度有限,给手术操作增加一定难度;(3)皮瓣供区无法直接缝合,往往需要皮片或皮瓣移植修复。以上不足在一定程度上限制了这个术式的广泛应用。
综上所述,本研究团队采用吻合皮神经的足底内侧游离皮瓣整复手掌电烧伤创面及瘢痕挛缩取得了较好的效果,值得临床推广。但本研究纳入的病例数有限,且为单中心回顾性研究,故仍需总结更多的临床病例,积累更多的临床经验从而进一步提高疗效。
孙礼祥:实验操作、数据整理、论文撰写、经费支持;吴帅:数据处理与论文撰写;张小薇:实验操作;刘文杰:论文修改;张凌娟:研究指导、论文修改、经费支持所有作者均声明不存在利益冲突调节性T细胞(Treg)可以通过多种机制来减轻炎症反应和防止自身免疫反应。常驻在外周组织(即非淋巴组织)的Treg有特异性的功能,比如,皮肤组织中的Treg可以促进创面愈合、抑制真皮纤维化、促进表皮再生和增强毛囊生长周期。皮肤组织中的Treg可通过促进影响上皮细胞生物学功能的整合素和TGF-β途径基因的表达,在转录水平上调整组织微环境。研究者观察到,小鼠表皮损伤后皮肤中的Treg促使KC发出急性促炎信号。使用单细胞测序方法,研究者确定了小鼠皮肤损伤时整合素αvβ8会在皮肤Treg上优先表达,Treg可以凭借该整合素激活潜伏的TGF-β,活化的TGF-β则可以直接作用于上皮细胞以促进趋化因子配体5的产生和中性粒细胞的募集。这一信号通路的激活抑制了小鼠表皮的再生,同时降低了金黄色葡萄球菌通过受损的皮肤屏障造成感染的概率。因此,皮肤中表达αvβ8的Treg与其典型的免疫抑制功能似乎存在矛盾,在皮肤屏障的完整性丧失后表现为促进炎症的急性发作,进而促进宿主对细菌感染的防御。张凡,编译自《Sci Immunol》, 2021,6(62):eabg2329;肖健,审校 -
参考文献
(37) [1] GalloRL,HooperLV.Epithelial antimicrobial defence of the skin and intestine[J].Nat Rev Immunol,2012,12(7):503-516.DOI: 10.1038/nri3228. [2] Cañedo-DorantesL,Cañedo-AyalaM.Skin acute wound healing: a comprehensive review[J].Int J Inflam,2019,2019:3706315.DOI: 10.1155/2019/3706315. [3] SorgH,TilkornDJ,HagerS,et al.Skin wound healing: an update on the current knowledge and concepts[J].Eur Surg Res,2017,58(1/2):81-94.DOI: 10.1159/000454919. [4] GrennanD.Diabetic foot ulcers[J].JAMA,2019,321(1):114.DOI: 10.1001/jama.2018.18323. [5] HanG,CeilleyR.Chronic wound healing: a review of current management and treatments[J].Adv Ther,2017,34(3):599-610.DOI: 10.1007/s12325-017-0478-y. [6] AlsterTS,TanziEL.Hypertrophic scars and keloids: etiology and management[J].Am J Clin Dermatol,2003,4(4):235-243.DOI: 10.2165/00128071-200304040-00003. [7] KischerCW,ThiesAC,ChvapilM.Perivascular myofibroblasts and microvascular occlusion in hypertrophic scars and keloids[J].Hum Pathol,1982,13(9):819-824.DOI: 10.1016/s0046-8177(82)80078-6. [8] BarrientosS,StojadinovicO,GolinkoMS,et al.Growth factors and cytokines in wound healing[J].Wound Repair Regen,2008,16(5):585-601.DOI: 10.1111/j.1524-475X.2008.00410.x. [9] AmiriN,GolinAP,JaliliRB,et al.Roles of cutaneous cell-cell communication in wound healing outcome: an emphasis on keratinocyte-fibroblast crosstalk[J].Exp Dermatol,2022,31(4):475-484.DOI: 10.1111/exd.14516. [10] HanCM,ChengB,WuP.Clinical guideline on topical growth factors for skin wounds[J/OL].Burns Trauma,2020,8:tkaa035[2022-02-15].https://pubmed.ncbi.nlm.nih.gov/33015207/.DOI: 10.1093/burnst/tkaa035. [11] ZubairM,AhmadJ.Role of growth factors and cytokines in diabetic foot ulcer healing: a detailed review[J].Rev Endocr Metab Disord,2019,20(2):207-217.DOI: 10.1007/s11154-019-09492-1. [12] WeiY,LiJ,HuangY,et al.The clinical effectiveness and safety of using epidermal growth factor, fibroblast growth factor and granulocyte-macrophage colony stimulating factor as therapeutics in acute skin wound healing: a systematic review and meta-analysis[J/OL].Burns Trauma,2022,10:tkac002[2022-02-15]. https://pubmed.ncbi.nlm.nih.gov/35265723/.DOI: 10.1093/burnst/tkac002. [13] ChenK,RaoZ,DongS,et al.Roles of the fibroblast growth factor signal transduction system in tissue injury repair[J/OL].Burns Trauma,2022,10:tkac005[2022-02-15]. https://pubmed.ncbi.nlm.nih.gov/35350443/.DOI: 10.1093/burnst/tkac005. [14] LiuY,LiuY,DengJ,et al.Fibroblast growth factor in diabetic foot ulcer: progress and therapeutic prospects[J].Front Endocrinol (Lausanne),2021,12:744868.DOI: 10.3389/fendo.2021.744868. [15] HuiQ,JinZ,LiX,et al.FGF family: from drug development to clinical application[J].Int J Mol Sci,2018,19(7):1875.DOI: 10.3390/ijms19071875. [16] KrookMA,ReeserJW,ErnstG,et al.Fibroblast growth factor receptors in cancer: genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance[J].Br J Cancer,2021,124(5):880-892.DOI: 10.1038/s41416-020-01157-0. [17] TracyLE,MinasianRA,CatersonEJ.Extracellular matrix and dermal fibroblast function in the healing wound[J].Adv Wound Care (New Rochelle),2016,5(3):119-136.DOI: 10.1089/wound.2014.0561. [18] desJardins-ParkHE,FosterDS,LongakerMT.Fibroblasts and wound healing: an update[J].Regen Med,2018,13(5):491-495.DOI: 10.2217/rme-2018-0073. [19] DriskellRR,LichtenbergerBM,HosteE,et al.Distinct fibroblast lineages determine dermal architecture in skin development and repair[J].Nature,2013,504(7479):277-281.DOI: 10.1038/nature12783. [20] TabibT,MorseC,WangT,et al.SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human skin[J].J Invest Dermatol,2018,138(4):802-810.DOI: 10.1016/j.jid.2017.09.045. [21] ZhangZ,ShaoM,HeplerC,et al.Dermal adipose tissue has high plasticity and undergoes reversible dedifferentiation in mice[J].J Clin Invest,2019,129(12):5327-5342.DOI: 10.1172/JCI130239. [22] HaenselD,JinS,SunP,et al.Defining epidermal basal cell states during skin homeostasis and wound healing using single-cell transcriptomics[J].Cell Rep,2020,30(11):3932-3947.e6.DOI: 10.1016/j.celrep.2020.02.091. [23] Guerrero-JuarezCF,DedhiaPH,JinS,et al.Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds[J].Nat Commun,2019,10(1):650.DOI: 10.1038/s41467-018-08247-x. [24] ZhangLJ,ChenSX,Guerrero-JuarezCF,et al.Age-related loss of innate immune antimicrobial function of dermal fat is mediated by transforming growth factor beta[J].Immunity,2019,50(1):121-136.e5.DOI: 10.1016/j.immuni.2018.11.003. [25] ZhangX,LanY,XuJ,et al.CellMarker: a manually curated resource of cell markers in human and mouse[J].Nucleic Acids Res,2019,47(D1):D721-D728.DOI: 10.1093/nar/gky900. [26] JinS,Guerrero-JuarezCF,ZhangL,et al.Inference and analysis of cell-cell communication using CellChat[J].Nat Commun,2021,12(1):1088.DOI: 10.1038/s41467-021-21246-9. [27] MerrickD,SakersA,IrgebayZ,et al.Identification of a mesenchymal progenitor cell hierarchy in adipose tissue[J].Science,2019,364(6438):eaav2501.DOI: 10.1126/science.aav2501. [28] Berry-KilgourC,CabralJ,WiseL.Advancements in the delivery of growth factors and cytokines for the treatment of cutaneous wound indications[J].Adv Wound Care (New Rochelle),2021,10(11):596-622.DOI: 10.1089/wound.2020.1183. [29] YamakawaS,HayashidaK.Advances in surgical applications of growth factors for wound healing[J/OL].Burns Trauma,2019,7:10[2022-02-15].https://pubmed.ncbi.nlm.nih.gov/30993143/.DOI: 10.1186/s41038-019-0148-1. [30] MaddalunoL,UrwylerC,WernerS.Fibroblast growth factors: key players in regeneration and tissue repair[J].Development,2017,144(22):4047-4060.DOI: 10.1242/dev.152587. [31] XieY,SuN,YangJ,et al.FGF/FGFR signaling in health and disease[J].Signal Transduct Target Ther,2020,5(1):181.DOI: 10.1038/s41392-020-00222-7. [32] LindnerV,MajackRA,ReidyMA.Basic fibroblast growth factor stimulates endothelial regrowth and proliferation in denuded arteries[J].J Clin Invest,1990,85(6):2004-2008.DOI: 10.1172/JCI114665. [33] TaiAL,ShamJS,XieD,et al.Co-overexpression of fibroblast growth factor 3 and epidermal growth factor receptor is correlated with the development of nonsmall cell lung carcinoma[J].Cancer,2006,106(1):146-155.DOI: 10.1002/cncr.21581. [34] HuL,ShamJS,XieD,et al.Up-regulation of fibroblast growth factor 3 is associated with tumor metastasis and recurrence in human hepatocellular carcinoma[J].Cancer Lett,2007,252(1):36-42.DOI: 10.1016/j.canlet.2006.12.003. [35] YenTT,ThaoDT,ThuocTL.An overview on keratinocyte growth factor: from the molecular properties to clinical applications[J].Protein Pept Lett,2014,21(3):306-317.DOI: 10.2174/09298665113206660115. [36] WernerS,KriegT,SmolaH.Keratinocyte-fibroblast interactions in wound healing[J].J Invest Dermatol,2007,127(5):998-1008.DOI: 10.1038/sj.jid.5700786. [37] ZinkleA,MohammadiM.Structural bology of the FGF7 subfamily[J].Front Genet,2019,10:102.DOI: 10.3389/fgene.2019.00102. -
3 基于健康小鼠正常皮肤组织和全层皮肤缺损小鼠伤后7 d创面组织细胞单细胞RNA测序数据的、表达血小板衍生生长因子受体α基因的、真皮成纤维细胞亚群的特定基因表达情况气泡图
注:从左至右各列分别对应细胞群C0、C5、C7、C1、C2、C4、C17、C6、C13;左侧缩写与中文均为基因名称,DPP4为二肽基肽酶4,LY6A为淋巴细胞抗原6A,ICAM1为细胞间黏附分子1,CRAMP为组织蛋白酶抑制素,PLIN2为脂滴包被蛋白2,CXCL为C-X-C基序趋化因子,IL-4为白细胞介素4,VCAM1为血管细胞黏附分子1,PCNA为增殖细胞核抗原,SAA3为血清淀粉样蛋白A-3,LGR5为富含亮氨酸重复的G蛋白偶联受体5,CSF1R为集落刺激因子1受体,COLⅠα1为Ⅰ型胶原α1链蛋白,ENTPD1为外核苷三磷酸二磷酸水解酶1,LRIG1为亮氨酸丰富重复免疫球蛋白样域蛋白1,LEF1为淋巴增强子结合因子1,ALPL为肝肾骨碱性磷酸酶,ACTα2为肌动蛋白α2,TAGLN为肌动蛋白凝胶蛋白,MYL1为肌球蛋白轻链1,MYLPF为肌球蛋白可磷酸化调节轻链
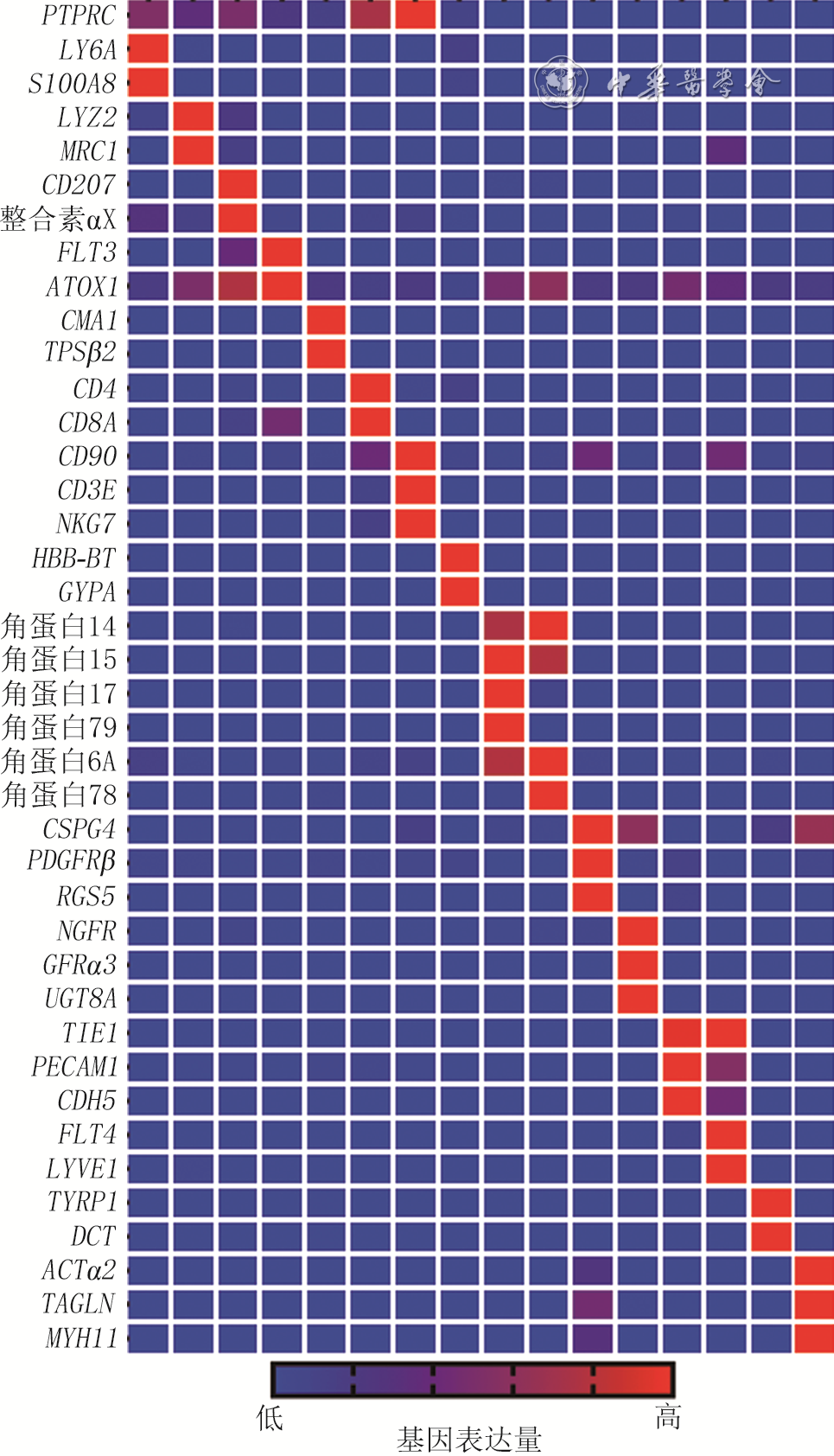
4 基于健康小鼠正常皮肤组织和全层皮肤缺损小鼠伤后7 d创面组织细胞单细胞RNA测序数据的、非真皮成纤维细胞群的特定基因表达情况热图
注:从左至右各列分别对应细胞群C3、C10、C12、C15、C22、C8、C20、C18、C9、C11、C14、C16、C19、C21、C23、C24;左侧缩写与中文均为基因名称,PTPRC为受体型蛋白酪氨酸磷酸酶C,LY6A为淋巴细胞抗原6A,S100A8为S100钙结合蛋白A8,MRC1为甘露糖受体C1,CMA1为糜蛋白酶1,TPSβ2为类胰蛋白酶β2,NKG为自然杀伤细胞相关蛋白,GYPA为血型糖蛋白A,CSPG4为硫酸软骨素蛋白聚糖4,PDGFRβ为血小板衍生生长因子受体β,RGS5为G蛋白信号通路调节蛋白5,NGFR为神经生长因子受体,GFRα3为胶质细胞源性神经营养因子家族受体α3,UGT8A为尿苷二磷酸糖基转移酶8A,TIE1为免疫球蛋白样表皮生长因子样域酪氨酸激酶1,PECAM1为血小板内皮细胞黏附分子1,LYVE1为淋巴管内皮透明质酸受体1,TYRP1为5,6-二羟基吲哚-2-羧酸氧化酶,DCT为多巴色素互变异构酶,ACTα2为肌动蛋白α2,TAGLN为肌动蛋白凝胶蛋白,MYH11为肌球蛋白轻链11
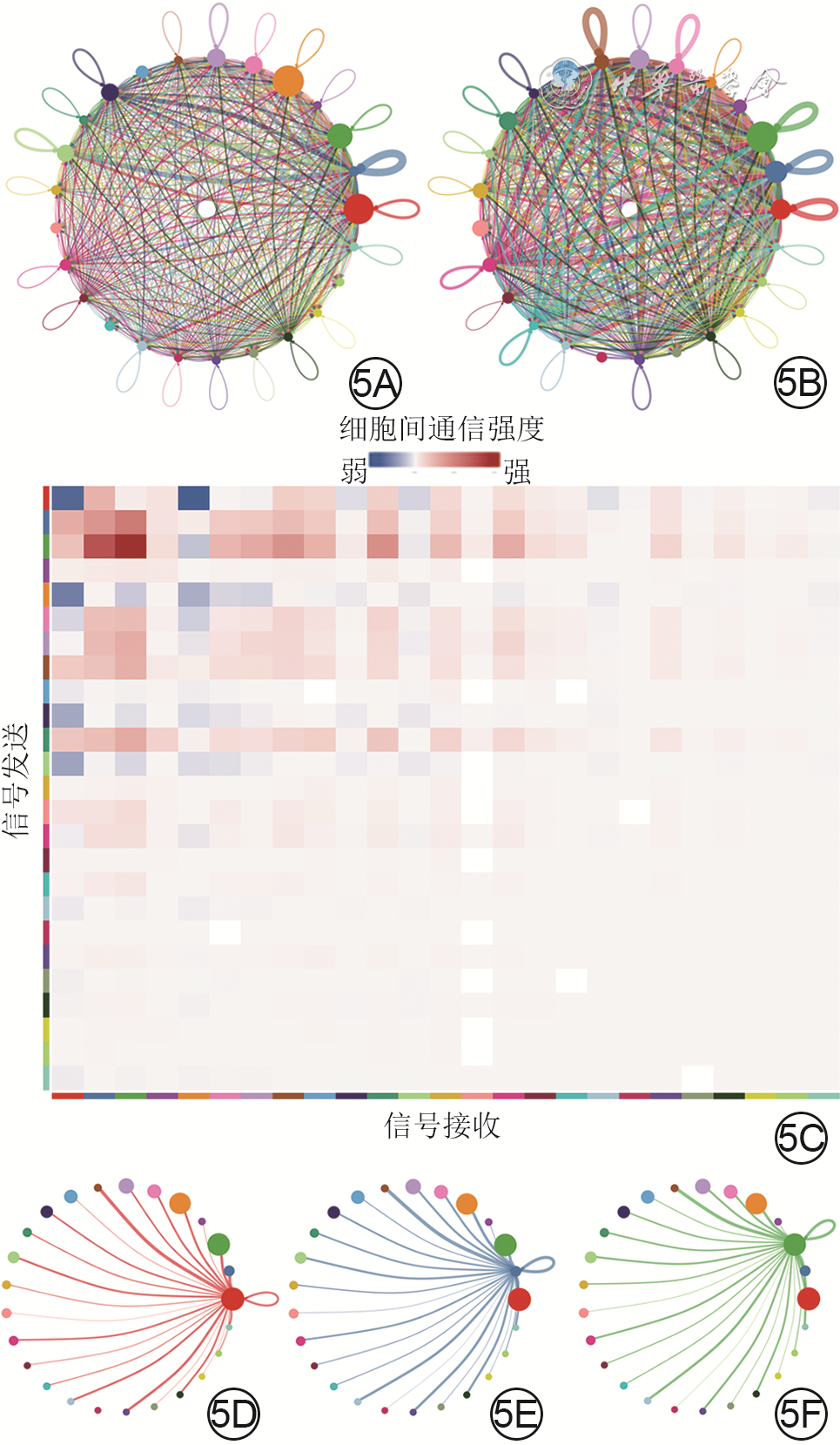
5 基于健康小鼠正常皮肤组织和全层皮肤缺损小鼠伤后7 d创面组织细胞单细胞RNA测序数据的细胞间通信网络图与热图。5A、5B.分别为正常皮肤组织、创面组织中细胞间通信情况网络图,图5B中细胞间通信较图5A更多更密集;5C.创面组织中细胞间通信强度热图;5D、5E、5F.分别为创面组织中细胞群C0、C1、C2与其他细胞群通信情况网络图
注:图5A、5B、5D、5E、5F中从最右侧红点开始,逆时针排列的25个点代表25个细胞群,对应编号为C0~C24,图中连线代表细胞间通信,连线的颜色与信号发送细胞群颜色一致,连线的粗细代表信号强度,点大小代表细胞群中细胞量;图5C中从左侧顶部/底部最左侧红色块开始,向下/右排列的25个色块代表25个细胞群,对应编号为C0~C24
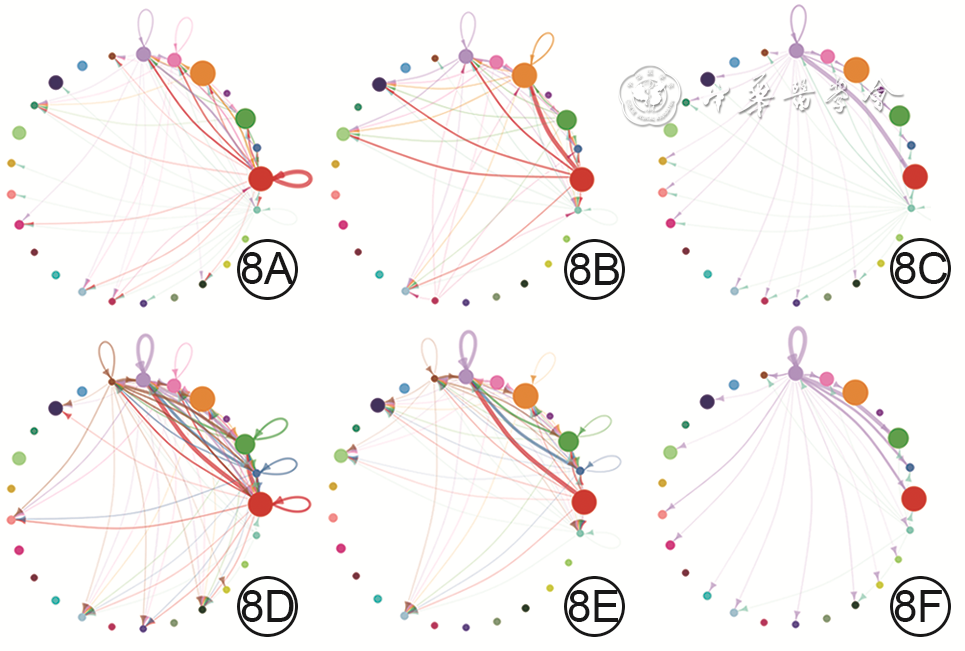
8 基于健康小鼠正常皮肤组织和全层皮肤缺损小鼠伤后7 d创面组织细胞单细胞RNA测序数据的、成纤维细胞生长因子(FGF)的各亚型与FGF受体(FGFR)各亚型的两两配对信号通路中的细胞间通信网络图。8A、8B、8C.分别为正常皮肤组织中FGF7-FGFR1、FGF7-FGFR2、FGF10-FGFR1信号通路中的细胞间通信,图8A中的细胞间通信较少,图8B中的细胞间通信较多,图8C中的细胞间通信少;8D、8E、8F.分别为创面组织中FGF7-FGFR1、FGF7-FGFR2、FGF10-FGFR1信号通路中的细胞间通信,图8D中细胞间通信明显多于图8A,图8E中细胞间通信稍少于图8B,图8F中细胞间通信与图8C相近,图8E、8F中细胞间通信明显少于图8D
注:网络图中从最右侧红点开始,逆时针排列的25个点代表25个细胞群,对应编号为C0~C24;网络图中连线代表细胞间通信,连线的颜色与信号发送细胞群颜色一致,连线的粗细代表信号强度,点大小代表细胞群中细胞量
9 健康小鼠正常皮肤组织和全层皮肤缺损小鼠伤后7 d创面组织中FGF7蛋白表达与分布及其与DPP4、SCA1、SMA、PDGFRα蛋白的共定位表达 花青素3-异硫氰酸荧光素-花青素5-4',6-二脒基-2-苯基吲哚×200,图中标尺为2 mm。9A、9B、9C、9D与9E、9F、9G、9H.分别为正常皮肤组织与创面组织中FGF7染色、DPP4染色、SCA1染色、FGF7与DPP4和SCA1及细胞核染色的重叠图片,图9A中FGF7蛋白表达量少,主要位于皮肤间质层且在靠近真皮白色脂肪组织附近也有表达,图9B中DPP4蛋白表达较多,图9C中SCA1蛋白表达较多,图9D中FGF7与DPP4和SCA1的蛋白共定位表达较少,图9E中FGF7蛋白表达量明显多于图9A,图9F中DPP4蛋白表达量与图9B相近,图9G中SCA1蛋白表达量与图9C相近,图9H中FGF7与DPP4和SCA1的蛋白共定位表达明显多于图9D;9I、9J、9K、9L与9M、9N、9O、9P.分别为正常皮肤组织与创面组织中FGF7染色、SMA染色、PDGFRα染色、FGF7与SMA和PDGFRα及细胞核染色的重叠图片,图9I中FGF7蛋白表达部位与图9A相近但量较少,图9J中SMA蛋白表达量少,图9K中PDGFRα蛋白表达量较少,图9L中FGF7蛋白与SMA蛋白无共定位且和PDGFRα蛋白的共定位表达少,图9M中FGF7蛋白表达量明显多于图9I,图9N中SMA蛋白表达明显多于图9J,图9O中PDGFRα蛋白表达与图9K相近,图9P中FGF7蛋白与SMA蛋白无共定位且与PDGFRα蛋白的共定位表达明显多于图9L
注:成纤维细胞生长因子7(FGF7)阳性染色为红色,二肽基肽酶4(DPP4)与平滑肌肌动蛋白(SMA)阳性染色均为绿色,干细胞抗原1 (SCA1)与血小板衍生生长因子受体α(PDGFRα)阳性染色均为蓝紫色,细胞核染色为白色;绿色箭头指示FGF7蛋白在间质层的分布,黄色箭头指示FGF7蛋白在白色脂肪组织附近的分布,红色箭头指示FGF7蛋白与其他蛋白共定位位置
-







 下载:
下载:



 下载:
下载: